library("yulab.utils")15 Gallery of Reproducible Examples
15.1 Visualizing pairwise nucleotide sequence distance with a phylogenetic tree
This example reproduces figure 1 of (Chen et al., 2017). It extracts accession numbers from tip labels of the HPV58 tree and calculates pairwise nucleotide sequence distances. The distance matrix is visualized as dot and line plots. This example demonstrates the ability to add multiple layers to a specific panel. As illustrated in Figure Figure 15.1, the geom_facet() function displays sequence distances as a dot plot and then adds a layer of line plot to the same panel, i.e., sequence distance. In addition, the tree in geom_facet() can be fully annotated with multiple layers (clade labels, bootstrap support values, etc.). The source code is modified from the supplemental file of (Yu et al., 2018).
library(TDbook)
library(tibble)
library(tidyr)
library(Biostrings)Loading required package: BiocGenerics
Attaching package: 'BiocGenerics'The following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, aperm, append, as.data.frame, basename, cbind,
colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
Position, rank, rbind, Reduce, rownames, sapply, saveRDS, setdiff,
table, tapply, union, unique, unsplit, which.max, which.minLoading required package: S4VectorsLoading required package: stats4
Attaching package: 'S4Vectors'The following object is masked from 'package:tidyr':
expandThe following object is masked from 'package:utils':
findMatchesThe following objects are masked from 'package:base':
expand.grid, I, unnameLoading required package: IRangesLoading required package: XVectorLoading required package: GenomeInfoDb
Attaching package: 'Biostrings'The following object is masked from 'package:base':
strsplitlibrary(treeio)treeio v1.35.0 Learn more at https://yulab-smu.top/contribution-tree-data/
Please cite:
LG Wang, TTY Lam, S Xu, Z Dai, L Zhou, T Feng, P Guo, CW Dunn, BR
Jones, T Bradley, H Zhu, Y Guan, Y Jiang, G Yu. treeio: an R package
for phylogenetic tree input and output with richly annotated and
associated data. Molecular Biology and Evolution. 2020, 37(2):599-603.
doi: 10.1093/molbev/msz240
Attaching package: 'treeio'The following object is masked from 'package:Biostrings':
masklibrary(ggplot2)
library(ggtree)ggtree v4.1.1.004 Learn more at https://yulab-smu.top/contribution-tree-data/
Please cite:
Shuangbin Xu, Lin Li, Xiao Luo, Meijun Chen, Wenli Tang, Li Zhan, Zehan
Dai, Tommy T. Lam, Yi Guan, Guangchuang Yu. Ggtree: A serialized data
object for visualization of a phylogenetic tree and annotation data.
iMeta 2022, 1(4):e56. doi:10.1002/imt2.56
Attaching package: 'ggtree'The following object is masked from 'package:Biostrings':
collapseThe following object is masked from 'package:IRanges':
collapseThe following object is masked from 'package:S4Vectors':
expandThe following object is masked from 'package:tidyr':
expand# loaded from TDbook package
tree <- tree_HPV58
clade <- c(A3 = 92, A1 = 94, A2 = 108, B1 = 156,
B2 = 159, C = 163, D1 = 173, D2 = 176)
tree <- groupClade(tree, clade)
cols <- c(A1 = "#EC762F", A2 = "#CA6629", A3 = "#894418", B1 = "#0923FA",
B2 = "#020D87", C = "#000000", D1 = "#9ACD32",D2 = "#08630A")
## visualize the tree with tip labels and tree scale
p <- ggtree(tree, aes(color = group), ladderize = FALSE) %>%
rotate(rootnode(tree)) +
geom_tiplab(aes(label = paste0("italic('", label, "')")),
parse = TRUE, size = 2.5) +
geom_treescale(x = 0, y = 1, width = 0.002) +
scale_color_manual(values = c(cols, "black"),
na.value = "black", name = "Lineage",
breaks = c("A1", "A2", "A3", "B1", "B2", "C", "D1", "D2")) +
guides(color = guide_legend(override.aes = list(size = 5, shape = 15))) +
theme_tree2(legend.position = c(.1, .88))Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
ℹ The deprecated feature was likely used in the ggtree package.
Please report the issue at <https://github.com/YuLab-SMU/ggtree/issues>.## Optional
## add labels for monophyletic (A, C and D) and paraphyletic (B) groups
dat <- tibble(node = c(94, 108, 131, 92, 156, 159, 163, 173, 176,172),
name = c("A1", "A2", "A3", "A", "B1",
"B2", "C", "D1", "D2", "D"),
offset = c(0.003, 0.003, 0.003, 0.00315, 0.003,
0.003, 0.0031, 0.003, 0.003, 0.00315),
offset.text = c(-.001, -.001, -.001, 0.0002, -.001,
-.001, 0.0002, -.001, -.001, 0.0002),
barsize = c(1.2, 1.2, 1.2, 2, 1.2, 1.2, 3.2, 1.2, 1.2, 2),
extend = list(c(0, 0.5), 0.5, c(0.5, 0), 0, c(0, 0.5),
c(0.5, 0), 0, c(0, 0.5), c(0.5, 0), 0)
) %>%
dplyr::group_split(barsize)
p <- p +
geom_cladelab(
data = dat[[1]],
mapping = aes(
node = node,
label = name,
color = group,
offset = offset,
offset.text = offset.text,
extend = extend
),
barsize = 1.2,
fontface = 3,
align = TRUE
) +
geom_cladelab(
data = dat[[2]],
mapping = aes(
node = node,
label = name,
offset = offset,
offset.text =offset.text,
extend = extend
),
barcolor = "darkgrey",
textcolor = "darkgrey",
barsize = 2,
fontsize = 5,
fontface = 3,
align = TRUE
) +
geom_cladelab(
data = dat[[3]],
mapping = aes(
node = node,
label = name,
offset = offset,
offset.text = offset.text,
extend = extend
),
barcolor = "darkgrey",
textcolor = "darkgrey",
barsize = 3.2,
fontsize = 5,
fontface = 3,
align = TRUE
) +
geom_strip(65, 71, "italic(B)", color = "darkgrey",
offset = 0.00315, align = TRUE, offset.text = 0.0002,
barsize = 2, fontsize = 5, parse = TRUE)
## Optional
## display support values
p <- p + geom_nodelab(aes(subset = (node == 92), label = "*"),
color = "black", nudge_x = -.001, nudge_y = 1) +
geom_nodelab(aes(subset = (node == 155), label = "*"),
color = "black", nudge_x = -.0003, nudge_y = -1) +
geom_nodelab(aes(subset = (node == 158), label = "95/92/1.00"),
color = "black", nudge_x = -0.0001,
nudge_y = -1, hjust = 1) +
geom_nodelab(aes(subset = (node == 162), label = "98/97/1.00"),
color = "black", nudge_x = -0.0001,
nudge_y = -1, hjust = 1) +
geom_nodelab(aes(subset = (node == 172), label = "*"),
color = "black", nudge_x = -.0003, nudge_y = -1) ## extract accession numbers from tip labels
tl <- tree$tip.label
acc <- sub("\\w+\\|", "", tl)
names(tl) <- acc
## read sequences from GenBank directly into R
tipseq <- seqmagick::ncbi_fa_read(acc)
## align the sequences using muscle
tipseq_aln <- muscle::muscle(tipseq)
MUSCLE v3.8.31 by Robert C. Edgar
http://www.drive5.com/muscle
This software is donated to the public domain.
Please cite: Edgar, R.C. Nucleic Acids Res 32(5), 1792-97.
file24cbd68f864 90 seqs, max length 7863, avg length 7826
1301 MB(8%)00:00:00 Iter 1 0.02% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 12.23% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 24.44% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 36.65% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 48.86% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 61.07% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 73.28% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 85.49% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 97.70% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 100.00% K-mer dist pass 1
1301 MB(8%)00:00:00 Iter 1 0.02% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 12.23% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 24.44% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 36.65% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 48.86% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 61.07% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 73.28% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 85.49% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 97.70% K-mer dist pass 2
1301 MB(8%)00:00:00 Iter 1 100.00% K-mer dist pass 2
1315 MB(8%)00:00:00 Iter 1 1.12% Align node
1391 MB(8%)00:00:01 Iter 1 2.25% Align node
1402 MB(9%)00:00:02 Iter 1 3.37% Align node
1405 MB(9%)00:00:03 Iter 1 4.49% Align node
1411 MB(9%)00:00:03 Iter 1 5.62% Align node
1418 MB(9%)00:00:04 Iter 1 6.74% Align node
1425 MB(9%)00:00:05 Iter 1 7.87% Align node
1432 MB(9%)00:00:05 Iter 1 8.99% Align node
1443 MB(9%)00:00:06 Iter 1 10.11% Align node
1450 MB(9%)00:00:07 Iter 1 11.24% Align node
1453 MB(9%)00:00:08 Iter 1 12.36% Align node
1455 MB(9%)00:00:08 Iter 1 13.48% Align node
1457 MB(9%)00:00:09 Iter 1 14.61% Align node
1460 MB(9%)00:00:10 Iter 1 15.73% Align node
1462 MB(9%)00:00:11 Iter 1 16.85% Align node
1464 MB(9%)00:00:11 Iter 1 17.98% Align node
1467 MB(9%)00:00:12 Iter 1 19.10% Align node
1469 MB(9%)00:00:13 Iter 1 20.22% Align node
1471 MB(9%)00:00:13 Iter 1 21.35% Align node
1474 MB(9%)00:00:14 Iter 1 22.47% Align node
1476 MB(9%)00:00:15 Iter 1 23.60% Align node
1478 MB(9%)00:00:16 Iter 1 24.72% Align node
1481 MB(9%)00:00:16 Iter 1 25.84% Align node
1492 MB(9%)00:00:17 Iter 1 26.97% Align node
1506 MB(9%)00:00:18 Iter 1 28.09% Align node
1508 MB(9%)00:00:18 Iter 1 29.21% Align node
1517 MB(9%)00:00:19 Iter 1 30.34% Align node
1520 MB(9%)00:00:20 Iter 1 31.46% Align node
1522 MB(9%)00:00:21 Iter 1 32.58% Align node
1524 MB(9%)00:00:21 Iter 1 33.71% Align node
1527 MB(9%)00:00:22 Iter 1 34.83% Align node
1529 MB(9%)00:00:23 Iter 1 35.96% Align node
1531 MB(9%)00:00:24 Iter 1 37.08% Align node
1547 MB(9%)00:00:24 Iter 1 38.20% Align node
1552 MB(9%)00:00:25 Iter 1 39.33% Align node
1559 MB(10%)00:00:26 Iter 1 40.45% Align node
1568 MB(10%)00:00:26 Iter 1 41.57% Align node
1572 MB(10%)00:00:27 Iter 1 42.70% Align node
1575 MB(10%)00:00:28 Iter 1 43.82% Align node
1577 MB(10%)00:00:29 Iter 1 44.94% Align node
1579 MB(10%)00:00:29 Iter 1 46.07% Align node
1582 MB(10%)00:00:30 Iter 1 47.19% Align node
1584 MB(10%)00:00:31 Iter 1 48.31% Align node
1587 MB(10%)00:00:32 Iter 1 49.44% Align node
1589 MB(10%)00:00:32 Iter 1 50.56% Align node
1591 MB(10%)00:00:33 Iter 1 51.69% Align node
1594 MB(10%)00:00:34 Iter 1 52.81% Align node
1600 MB(10%)00:00:34 Iter 1 53.93% Align node
1619 MB(10%)00:00:35 Iter 1 55.06% Align node
1621 MB(10%)00:00:36 Iter 1 56.18% Align node
1623 MB(10%)00:00:37 Iter 1 57.30% Align node
1626 MB(10%)00:00:37 Iter 1 58.43% Align node
1628 MB(10%)00:00:38 Iter 1 59.55% Align node
1630 MB(10%)00:00:39 Iter 1 60.67% Align node
1646 MB(10%)00:00:40 Iter 1 61.80% Align node
1649 MB(10%)00:00:40 Iter 1 62.92% Align node
1651 MB(10%)00:00:41 Iter 1 64.04% Align node
1653 MB(10%)00:00:42 Iter 1 65.17% Align node
1656 MB(10%)00:00:42 Iter 1 66.29% Align node
1658 MB(10%)00:00:43 Iter 1 67.42% Align node
1660 MB(10%)00:00:44 Iter 1 68.54% Align node
1663 MB(10%)00:00:45 Iter 1 69.66% Align node
1665 MB(10%)00:00:45 Iter 1 70.79% Align node
1679 MB(10%)00:00:46 Iter 1 71.91% Align node
1681 MB(10%)00:00:47 Iter 1 73.03% Align node
1683 MB(10%)00:00:48 Iter 1 74.16% Align node
1686 MB(10%)00:00:48 Iter 1 75.28% Align node
1697 MB(10%)00:00:49 Iter 1 76.40% Align node
1699 MB(10%)00:00:50 Iter 1 77.53% Align node
1702 MB(10%)00:00:50 Iter 1 78.65% Align node
1704 MB(10%)00:00:51 Iter 1 79.78% Align node
1713 MB(10%)00:00:52 Iter 1 80.90% Align node
1715 MB(10%)00:00:53 Iter 1 82.02% Align node
1725 MB(11%)00:00:53 Iter 1 83.15% Align node
1736 MB(11%)00:00:54 Iter 1 84.27% Align node
1738 MB(11%)00:00:55 Iter 1 85.39% Align node
1741 MB(11%)00:00:56 Iter 1 86.52% Align node
1743 MB(11%)00:00:56 Iter 1 87.64% Align node
1745 MB(11%)00:00:57 Iter 1 88.76% Align node
1748 MB(11%)00:00:58 Iter 1 89.89% Align node
1757 MB(11%)00:00:58 Iter 1 91.01% Align node
1759 MB(11%)00:00:59 Iter 1 92.13% Align node
1766 MB(11%)00:01:00 Iter 1 93.26% Align node
1773 MB(11%)00:01:01 Iter 1 94.38% Align node
1775 MB(11%)00:01:01 Iter 1 95.51% Align node
1778 MB(11%)00:01:02 Iter 1 96.63% Align node
1780 MB(11%)00:01:03 Iter 1 97.75% Align node
1782 MB(11%)00:01:04 Iter 1 98.88% Align node
1785 MB(11%)00:01:04 Iter 1 100.00% Align node
1787 MB(11%)00:01:05 Iter 1 100.00% Align node
1787 MB(11%)00:01:05 Iter 1 1.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 2.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 3.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 4.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 5.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 6.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 7.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 8.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 10.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 11.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 12.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 13.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 14.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 15.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 16.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 17.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 18.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 20.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 21.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 22.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 23.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 24.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 25.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 26.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 27.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 28.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 30.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 31.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 32.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 33.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 34.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 35.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 36.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 37.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 38.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 40.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 41.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 42.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 43.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 44.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 45.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 46.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 47.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 48.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 50.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 51.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 52.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 53.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 54.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 55.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 56.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 57.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 58.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 60.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 61.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 62.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 63.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 64.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 65.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 66.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 67.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 68.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 70.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 71.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 72.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 73.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 74.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 75.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 76.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 77.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 78.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 80.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 81.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 82.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 83.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 84.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 85.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 86.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 87.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 88.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 90.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 91.11% Root alignment
1788 MB(11%)00:01:05 Iter 1 92.22% Root alignment
1788 MB(11%)00:01:05 Iter 1 93.33% Root alignment
1788 MB(11%)00:01:05 Iter 1 94.44% Root alignment
1788 MB(11%)00:01:05 Iter 1 95.56% Root alignment
1788 MB(11%)00:01:05 Iter 1 96.67% Root alignment
1788 MB(11%)00:01:05 Iter 1 97.78% Root alignment
1788 MB(11%)00:01:05 Iter 1 98.89% Root alignment
1788 MB(11%)00:01:05 Iter 1 100.00% Root alignment
1788 MB(11%)00:01:05 Iter 1 100.00% Root alignment
1788 MB(11%)00:01:05 Iter 2 1.14% Refine tree
1790 MB(11%)00:01:06 Iter 2 2.27% Refine tree
1790 MB(11%)00:01:07 Iter 2 3.41% Refine tree
1790 MB(11%)00:01:07 Iter 2 4.55% Refine tree
1790 MB(11%)00:01:08 Iter 2 5.68% Refine tree
1790 MB(11%)00:01:09 Iter 2 6.82% Refine tree
1790 MB(11%)00:01:10 Iter 2 7.95% Refine tree
1790 MB(11%)00:01:10 Iter 2 9.09% Refine tree
1790 MB(11%)00:01:11 Iter 2 10.23% Refine tree
1790 MB(11%)00:01:12 Iter 2 11.36% Refine tree
1790 MB(11%)00:01:12 Iter 2 12.50% Refine tree
1790 MB(11%)00:01:13 Iter 2 13.64% Refine tree
1790 MB(11%)00:01:14 Iter 2 14.77% Refine tree
1790 MB(11%)00:01:15 Iter 2 15.91% Refine tree
1790 MB(11%)00:01:15 Iter 2 17.05% Refine tree
1790 MB(11%)00:01:16 Iter 2 18.18% Refine tree
1790 MB(11%)00:01:17 Iter 2 19.32% Refine tree
1790 MB(11%)00:01:17 Iter 2 20.45% Refine tree
1790 MB(11%)00:01:18 Iter 2 21.59% Refine tree
1790 MB(11%)00:01:19 Iter 2 22.73% Refine tree
1790 MB(11%)00:01:20 Iter 2 23.86% Refine tree
1790 MB(11%)00:01:20 Iter 2 25.00% Refine tree
1790 MB(11%)00:01:21 Iter 2 26.14% Refine tree
1790 MB(11%)00:01:22 Iter 2 27.27% Refine tree
1790 MB(11%)00:01:23 Iter 2 28.41% Refine tree
1790 MB(11%)00:01:23 Iter 2 29.55% Refine tree
1790 MB(11%)00:01:24 Iter 2 30.68% Refine tree
1790 MB(11%)00:01:25 Iter 2 31.82% Refine tree
1790 MB(11%)00:01:25 Iter 2 32.95% Refine tree
1790 MB(11%)00:01:26 Iter 2 34.09% Refine tree
1790 MB(11%)00:01:27 Iter 2 35.23% Refine tree
1790 MB(11%)00:01:28 Iter 2 36.36% Refine tree
1790 MB(11%)00:01:28 Iter 2 37.50% Refine tree
1790 MB(11%)00:01:29 Iter 2 38.64% Refine tree
1790 MB(11%)00:01:30 Iter 2 39.77% Refine tree
1790 MB(11%)00:01:31 Iter 2 40.91% Refine tree
1790 MB(11%)00:01:31 Iter 2 42.05% Refine tree
1790 MB(11%)00:01:32 Iter 2 43.18% Refine tree
1790 MB(11%)00:01:33 Iter 2 44.32% Refine tree
1790 MB(11%)00:01:33 Iter 2 45.45% Refine tree
1790 MB(11%)00:01:34 Iter 2 46.59% Refine tree
1790 MB(11%)00:01:35 Iter 2 47.73% Refine tree
1790 MB(11%)00:01:36 Iter 2 48.86% Refine tree
1790 MB(11%)00:01:36 Iter 2 50.00% Refine tree
1790 MB(11%)00:01:37 Iter 2 51.14% Refine tree
1790 MB(11%)00:01:38 Iter 2 52.27% Refine tree
1790 MB(11%)00:01:39 Iter 2 53.41% Refine tree
1790 MB(11%)00:01:39 Iter 2 54.55% Refine tree
1790 MB(11%)00:01:40 Iter 2 100.00% Refine tree
1790 MB(11%)00:01:40 Iter 2 1.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 2.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 3.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 4.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 5.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 6.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 7.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 8.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 10.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 11.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 12.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 13.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 14.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 15.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 16.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 17.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 18.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 20.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 21.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 22.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 23.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 24.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 25.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 26.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 27.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 28.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 30.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 31.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 32.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 33.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 34.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 35.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 36.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 37.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 38.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 40.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 41.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 42.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 43.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 44.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 45.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 46.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 47.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 48.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 50.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 51.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 52.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 53.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 54.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 55.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 56.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 57.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 58.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 60.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 61.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 62.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 63.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 64.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 65.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 66.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 67.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 68.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 70.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 71.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 72.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 73.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 74.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 75.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 76.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 77.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 78.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 80.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 81.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 82.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 83.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 84.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 85.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 86.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 87.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 88.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 90.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 91.11% Root alignment
1790 MB(11%)00:01:40 Iter 2 92.22% Root alignment
1790 MB(11%)00:01:40 Iter 2 93.33% Root alignment
1790 MB(11%)00:01:40 Iter 2 94.44% Root alignment
1790 MB(11%)00:01:40 Iter 2 95.56% Root alignment
1790 MB(11%)00:01:40 Iter 2 96.67% Root alignment
1790 MB(11%)00:01:40 Iter 2 97.78% Root alignment
1790 MB(11%)00:01:40 Iter 2 98.89% Root alignment
1790 MB(11%)00:01:40 Iter 2 100.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 100.00% Root alignment
1790 MB(11%)00:01:40 Iter 2 100.00% Root alignment
1790 MB(11%)00:01:41 Iter 3 1.13% Refine biparts
1790 MB(11%)00:01:42 Iter 3 1.69% Refine biparts
1790 MB(11%)00:01:42 Iter 3 2.26% Refine biparts
1790 MB(11%)00:01:43 Iter 3 2.82% Refine biparts
1790 MB(11%)00:01:44 Iter 3 3.39% Refine biparts
1790 MB(11%)00:01:45 Iter 3 3.95% Refine biparts
1790 MB(11%)00:01:45 Iter 3 4.52% Refine biparts
1790 MB(11%)00:01:46 Iter 3 5.08% Refine biparts
1790 MB(11%)00:01:47 Iter 3 5.65% Refine biparts
1790 MB(11%)00:01:48 Iter 3 6.21% Refine biparts
1790 MB(11%)00:01:48 Iter 3 6.78% Refine biparts
1790 MB(11%)00:01:49 Iter 3 7.34% Refine biparts
1790 MB(11%)00:01:50 Iter 3 7.91% Refine biparts
1790 MB(11%)00:01:51 Iter 3 8.47% Refine biparts
1790 MB(11%)00:01:51 Iter 3 9.04% Refine biparts
1790 MB(11%)00:01:52 Iter 3 9.60% Refine biparts
1790 MB(11%)00:01:53 Iter 3 10.17% Refine biparts
1790 MB(11%)00:01:54 Iter 3 10.73% Refine biparts
1790 MB(11%)00:01:54 Iter 3 11.30% Refine biparts
1790 MB(11%)00:01:55 Iter 3 11.86% Refine biparts
1790 MB(11%)00:01:56 Iter 3 12.43% Refine biparts
1790 MB(11%)00:01:57 Iter 3 12.99% Refine biparts
1790 MB(11%)00:01:58 Iter 3 13.56% Refine biparts
1790 MB(11%)00:01:58 Iter 3 14.12% Refine biparts
1790 MB(11%)00:01:59 Iter 3 14.69% Refine biparts
1790 MB(11%)00:02:00 Iter 3 15.25% Refine biparts
1790 MB(11%)00:02:01 Iter 3 15.82% Refine biparts
1790 MB(11%)00:02:01 Iter 3 16.38% Refine biparts
1790 MB(11%)00:02:02 Iter 3 16.95% Refine biparts
1790 MB(11%)00:02:03 Iter 3 17.51% Refine biparts
1790 MB(11%)00:02:04 Iter 3 18.08% Refine biparts
1790 MB(11%)00:02:04 Iter 3 18.64% Refine biparts
1790 MB(11%)00:02:05 Iter 3 19.21% Refine biparts
1790 MB(11%)00:02:06 Iter 3 19.77% Refine biparts
1790 MB(11%)00:02:07 Iter 3 20.34% Refine biparts
1790 MB(11%)00:02:07 Iter 3 20.90% Refine biparts
1790 MB(11%)00:02:08 Iter 3 21.47% Refine biparts
1790 MB(11%)00:02:09 Iter 3 22.03% Refine biparts
1790 MB(11%)00:02:10 Iter 3 22.60% Refine biparts
1790 MB(11%)00:02:10 Iter 3 23.16% Refine biparts
1790 MB(11%)00:02:11 Iter 3 23.73% Refine biparts
1790 MB(11%)00:02:12 Iter 3 24.29% Refine biparts
1790 MB(11%)00:02:13 Iter 3 24.86% Refine biparts
1790 MB(11%)00:02:13 Iter 3 25.42% Refine biparts
1790 MB(11%)00:02:14 Iter 3 25.99% Refine biparts
1790 MB(11%)00:02:15 Iter 3 26.55% Refine biparts
1790 MB(11%)00:02:16 Iter 3 27.12% Refine biparts
1790 MB(11%)00:02:17 Iter 3 27.68% Refine biparts
1790 MB(11%)00:02:17 Iter 3 28.25% Refine biparts
1790 MB(11%)00:02:18 Iter 3 28.81% Refine biparts
1790 MB(11%)00:02:19 Iter 3 29.38% Refine biparts
1790 MB(11%)00:02:20 Iter 3 29.94% Refine biparts
1790 MB(11%)00:02:20 Iter 3 30.51% Refine biparts
1790 MB(11%)00:02:21 Iter 3 31.07% Refine biparts
1790 MB(11%)00:02:22 Iter 3 31.64% Refine biparts
1790 MB(11%)00:02:23 Iter 3 32.20% Refine biparts
1790 MB(11%)00:02:23 Iter 3 32.77% Refine biparts
1790 MB(11%)00:02:24 Iter 3 33.33% Refine biparts
1790 MB(11%)00:02:25 Iter 3 33.90% Refine biparts
1790 MB(11%)00:02:26 Iter 3 34.46% Refine biparts
1790 MB(11%)00:02:26 Iter 3 35.03% Refine biparts
1790 MB(11%)00:02:27 Iter 3 35.59% Refine biparts
1790 MB(11%)00:02:28 Iter 3 36.16% Refine biparts
1790 MB(11%)00:02:29 Iter 3 36.72% Refine biparts
1790 MB(11%)00:02:29 Iter 3 37.29% Refine biparts
1790 MB(11%)00:02:30 Iter 3 37.85% Refine biparts
1790 MB(11%)00:02:31 Iter 3 38.42% Refine biparts
1790 MB(11%)00:02:32 Iter 3 38.98% Refine biparts
1790 MB(11%)00:02:33 Iter 3 39.55% Refine biparts
1790 MB(11%)00:02:33 Iter 3 40.11% Refine biparts
1790 MB(11%)00:02:34 Iter 3 40.68% Refine biparts
1790 MB(11%)00:02:35 Iter 3 41.24% Refine biparts
1790 MB(11%)00:02:36 Iter 3 41.81% Refine biparts
1790 MB(11%)00:02:36 Iter 3 42.37% Refine biparts
1790 MB(11%)00:02:37 Iter 3 42.94% Refine biparts
1790 MB(11%)00:02:38 Iter 3 43.50% Refine biparts
1790 MB(11%)00:02:39 Iter 3 44.07% Refine biparts
1790 MB(11%)00:02:39 Iter 3 44.63% Refine biparts
1790 MB(11%)00:02:40 Iter 3 45.20% Refine biparts
1790 MB(11%)00:02:41 Iter 3 45.76% Refine biparts
1790 MB(11%)00:02:42 Iter 3 46.33% Refine biparts
1790 MB(11%)00:02:43 Iter 3 46.89% Refine biparts
1790 MB(11%)00:02:43 Iter 3 47.46% Refine biparts
1790 MB(11%)00:02:44 Iter 3 48.02% Refine biparts
1790 MB(11%)00:02:45 Iter 3 48.59% Refine biparts
1790 MB(11%)00:02:46 Iter 3 49.15% Refine biparts
1790 MB(11%)00:02:46 Iter 3 49.72% Refine biparts
1790 MB(11%)00:02:47 Iter 3 50.28% Refine biparts
1790 MB(11%)00:02:48 Iter 3 50.85% Refine biparts
1790 MB(11%)00:02:49 Iter 3 51.41% Refine biparts
1790 MB(11%)00:02:49 Iter 3 51.98% Refine biparts
1790 MB(11%)00:02:50 Iter 3 52.54% Refine biparts
1790 MB(11%)00:02:51 Iter 3 53.11% Refine biparts
1790 MB(11%)00:02:52 Iter 3 53.67% Refine biparts
1790 MB(11%)00:02:53 Iter 3 54.24% Refine biparts
1790 MB(11%)00:02:53 Iter 3 54.80% Refine biparts
1790 MB(11%)00:02:54 Iter 3 55.37% Refine biparts
1790 MB(11%)00:02:55 Iter 3 55.93% Refine biparts
1790 MB(11%)00:02:56 Iter 3 56.50% Refine biparts
1790 MB(11%)00:02:56 Iter 3 57.06% Refine biparts
1790 MB(11%)00:02:57 Iter 3 57.63% Refine biparts
1790 MB(11%)00:02:58 Iter 3 58.19% Refine biparts
1790 MB(11%)00:02:59 Iter 3 58.76% Refine biparts
1790 MB(11%)00:02:59 Iter 3 59.32% Refine biparts
1790 MB(11%)00:03:00 Iter 3 59.89% Refine biparts
1790 MB(11%)00:03:01 Iter 3 60.45% Refine biparts
1790 MB(11%)00:03:02 Iter 3 61.02% Refine biparts
1790 MB(11%)00:03:02 Iter 3 61.58% Refine biparts
1790 MB(11%)00:03:03 Iter 3 62.15% Refine biparts
1790 MB(11%)00:03:04 Iter 3 62.71% Refine biparts
1790 MB(11%)00:03:05 Iter 3 63.28% Refine biparts
1790 MB(11%)00:03:05 Iter 3 63.84% Refine biparts
1790 MB(11%)00:03:06 Iter 3 64.41% Refine biparts
1790 MB(11%)00:03:07 Iter 3 64.97% Refine biparts
1790 MB(11%)00:03:08 Iter 3 65.54% Refine biparts
1790 MB(11%)00:03:08 Iter 3 66.10% Refine biparts
1790 MB(11%)00:03:09 Iter 3 66.67% Refine biparts
1790 MB(11%)00:03:10 Iter 3 67.23% Refine biparts
1790 MB(11%)00:03:11 Iter 3 67.80% Refine biparts
1790 MB(11%)00:03:12 Iter 3 68.36% Refine biparts
1790 MB(11%)00:03:12 Iter 3 68.93% Refine biparts
1790 MB(11%)00:03:13 Iter 3 69.49% Refine biparts
1790 MB(11%)00:03:14 Iter 3 70.06% Refine biparts
1790 MB(11%)00:03:15 Iter 3 70.62% Refine biparts
1790 MB(11%)00:03:15 Iter 3 71.19% Refine biparts
1790 MB(11%)00:03:16 Iter 3 71.75% Refine biparts
1790 MB(11%)00:03:17 Iter 3 72.32% Refine biparts
1790 MB(11%)00:03:18 Iter 3 72.88% Refine biparts
1790 MB(11%)00:03:18 Iter 3 73.45% Refine biparts
1790 MB(11%)00:03:19 Iter 3 74.01% Refine biparts
1790 MB(11%)00:03:20 Iter 3 74.58% Refine biparts
1790 MB(11%)00:03:21 Iter 3 75.14% Refine biparts
1790 MB(11%)00:03:21 Iter 3 75.71% Refine biparts
1790 MB(11%)00:03:22 Iter 3 76.27% Refine biparts
1790 MB(11%)00:03:23 Iter 3 76.84% Refine biparts
1790 MB(11%)00:03:24 Iter 3 77.40% Refine biparts
1790 MB(11%)00:03:24 Iter 3 77.97% Refine biparts
1790 MB(11%)00:03:25 Iter 3 78.53% Refine biparts
1790 MB(11%)00:03:26 Iter 3 79.10% Refine biparts
1790 MB(11%)00:03:27 Iter 3 79.66% Refine biparts
1790 MB(11%)00:03:27 Iter 3 80.23% Refine biparts
1790 MB(11%)00:03:28 Iter 3 80.79% Refine biparts
1790 MB(11%)00:03:29 Iter 3 81.36% Refine biparts
1790 MB(11%)00:03:30 Iter 3 81.92% Refine biparts
1790 MB(11%)00:03:31 Iter 3 82.49% Refine biparts
1790 MB(11%)00:03:31 Iter 3 83.05% Refine biparts
1790 MB(11%)00:03:32 Iter 3 83.62% Refine biparts
1790 MB(11%)00:03:33 Iter 3 84.18% Refine biparts
1790 MB(11%)00:03:34 Iter 3 84.75% Refine biparts
1790 MB(11%)00:03:34 Iter 3 85.31% Refine biparts
1790 MB(11%)00:03:35 Iter 3 85.88% Refine biparts
1790 MB(11%)00:03:36 Iter 3 86.44% Refine biparts
1790 MB(11%)00:03:37 Iter 3 87.01% Refine biparts
1790 MB(11%)00:03:37 Iter 3 87.57% Refine biparts
1790 MB(11%)00:03:38 Iter 3 88.14% Refine biparts
1790 MB(11%)00:03:39 Iter 3 88.70% Refine biparts
1790 MB(11%)00:03:40 Iter 3 89.27% Refine biparts
1790 MB(11%)00:03:40 Iter 3 89.83% Refine biparts
1790 MB(11%)00:03:41 Iter 3 90.40% Refine biparts
1790 MB(11%)00:03:42 Iter 3 90.96% Refine biparts
1790 MB(11%)00:03:43 Iter 3 91.53% Refine biparts
1790 MB(11%)00:03:43 Iter 3 92.09% Refine biparts
1790 MB(11%)00:03:44 Iter 3 92.66% Refine biparts
1790 MB(11%)00:03:45 Iter 3 93.22% Refine biparts
1790 MB(11%)00:03:46 Iter 3 93.79% Refine biparts
1790 MB(11%)00:03:47 Iter 3 94.35% Refine biparts
1790 MB(11%)00:03:47 Iter 3 94.92% Refine biparts
1790 MB(11%)00:03:48 Iter 3 95.48% Refine biparts
1790 MB(11%)00:03:49 Iter 3 96.05% Refine biparts
1790 MB(11%)00:03:50 Iter 3 96.61% Refine biparts
1790 MB(11%)00:03:50 Iter 3 97.18% Refine biparts
1790 MB(11%)00:03:51 Iter 3 97.74% Refine biparts
1790 MB(11%)00:03:52 Iter 3 98.31% Refine biparts
1790 MB(11%)00:03:53 Iter 3 98.87% Refine biparts
1790 MB(11%)00:03:53 Iter 3 99.44% Refine biparts
1790 MB(11%)00:03:54 Iter 3 100.00% Refine biparts
1790 MB(11%)00:03:55 Iter 3 100.56% Refine biparts
1790 MB(11%)00:03:55 Iter 3 100.00% Refine biparts
1790 MB(11%)00:03:56 Iter 4 1.13% Refine biparts
1790 MB(11%)00:03:57 Iter 4 1.69% Refine biparts
1790 MB(11%)00:03:57 Iter 4 2.26% Refine biparts
1790 MB(11%)00:03:58 Iter 4 2.82% Refine biparts
1790 MB(11%)00:03:59 Iter 4 3.39% Refine biparts
1790 MB(11%)00:04:00 Iter 4 3.95% Refine biparts
1790 MB(11%)00:04:00 Iter 4 4.52% Refine biparts
1790 MB(11%)00:04:01 Iter 4 5.08% Refine biparts
1790 MB(11%)00:04:02 Iter 4 5.65% Refine biparts
1790 MB(11%)00:04:03 Iter 4 6.21% Refine biparts
1790 MB(11%)00:04:03 Iter 4 6.78% Refine biparts
1790 MB(11%)00:04:04 Iter 4 7.34% Refine biparts
1790 MB(11%)00:04:05 Iter 4 7.91% Refine biparts
1790 MB(11%)00:04:06 Iter 4 8.47% Refine biparts
1790 MB(11%)00:04:06 Iter 4 9.04% Refine biparts
1790 MB(11%)00:04:07 Iter 4 9.60% Refine biparts
1790 MB(11%)00:04:08 Iter 4 10.17% Refine biparts
1790 MB(11%)00:04:09 Iter 4 10.73% Refine biparts
1790 MB(11%)00:04:09 Iter 4 11.30% Refine biparts
1790 MB(11%)00:04:10 Iter 4 11.86% Refine biparts
1790 MB(11%)00:04:11 Iter 4 12.43% Refine biparts
1790 MB(11%)00:04:12 Iter 4 12.99% Refine biparts
1790 MB(11%)00:04:13 Iter 4 13.56% Refine biparts
1790 MB(11%)00:04:13 Iter 4 14.12% Refine biparts
1790 MB(11%)00:04:14 Iter 4 14.69% Refine biparts
1790 MB(11%)00:04:15 Iter 4 15.25% Refine biparts
1790 MB(11%)00:04:16 Iter 4 15.82% Refine biparts
1790 MB(11%)00:04:16 Iter 4 16.38% Refine biparts
1790 MB(11%)00:04:17 Iter 4 16.95% Refine biparts
1790 MB(11%)00:04:18 Iter 4 17.51% Refine biparts
1790 MB(11%)00:04:19 Iter 4 18.08% Refine biparts
1790 MB(11%)00:04:19 Iter 4 18.64% Refine biparts
1790 MB(11%)00:04:20 Iter 4 19.21% Refine biparts
1790 MB(11%)00:04:21 Iter 4 19.77% Refine biparts
1790 MB(11%)00:04:22 Iter 4 20.34% Refine biparts
1790 MB(11%)00:04:22 Iter 4 20.90% Refine biparts
1790 MB(11%)00:04:23 Iter 4 21.47% Refine biparts
1790 MB(11%)00:04:24 Iter 4 22.03% Refine biparts
1790 MB(11%)00:04:25 Iter 4 22.60% Refine biparts
1790 MB(11%)00:04:25 Iter 4 23.16% Refine biparts
1790 MB(11%)00:04:26 Iter 4 23.73% Refine biparts
1790 MB(11%)00:04:27 Iter 4 24.29% Refine biparts
1790 MB(11%)00:04:28 Iter 4 24.86% Refine biparts
1790 MB(11%)00:04:28 Iter 4 25.42% Refine biparts
1790 MB(11%)00:04:29 Iter 4 25.99% Refine biparts
1790 MB(11%)00:04:30 Iter 4 26.55% Refine biparts
1790 MB(11%)00:04:31 Iter 4 27.12% Refine biparts
1790 MB(11%)00:04:32 Iter 4 27.68% Refine biparts
1790 MB(11%)00:04:32 Iter 4 28.25% Refine biparts
1790 MB(11%)00:04:33 Iter 4 28.81% Refine biparts
1790 MB(11%)00:04:34 Iter 4 29.38% Refine biparts
1790 MB(11%)00:04:35 Iter 4 29.94% Refine biparts
1790 MB(11%)00:04:35 Iter 4 30.51% Refine biparts
1790 MB(11%)00:04:36 Iter 4 31.07% Refine biparts
1790 MB(11%)00:04:37 Iter 4 31.64% Refine biparts
1790 MB(11%)00:04:38 Iter 4 32.20% Refine biparts
1790 MB(11%)00:04:38 Iter 4 32.77% Refine biparts
1790 MB(11%)00:04:39 Iter 4 33.33% Refine biparts
1790 MB(11%)00:04:40 Iter 4 33.90% Refine biparts
1790 MB(11%)00:04:41 Iter 4 34.46% Refine biparts
1790 MB(11%)00:04:41 Iter 4 35.03% Refine biparts
1790 MB(11%)00:04:42 Iter 4 35.59% Refine biparts
1790 MB(11%)00:04:43 Iter 4 36.16% Refine biparts
1790 MB(11%)00:04:44 Iter 4 36.72% Refine biparts
1790 MB(11%)00:04:44 Iter 4 37.29% Refine biparts
1790 MB(11%)00:04:45 Iter 4 37.85% Refine biparts
1790 MB(11%)00:04:46 Iter 4 38.42% Refine biparts
1790 MB(11%)00:04:47 Iter 4 38.98% Refine biparts
1790 MB(11%)00:04:47 Iter 4 39.55% Refine biparts
1790 MB(11%)00:04:48 Iter 4 40.11% Refine biparts
1790 MB(11%)00:04:49 Iter 4 40.68% Refine biparts
1790 MB(11%)00:04:50 Iter 4 41.24% Refine biparts
1790 MB(11%)00:04:51 Iter 4 41.81% Refine biparts
1790 MB(11%)00:04:51 Iter 4 42.37% Refine biparts
1790 MB(11%)00:04:52 Iter 4 42.94% Refine biparts
1790 MB(11%)00:04:53 Iter 4 43.50% Refine biparts
1790 MB(11%)00:04:54 Iter 4 44.07% Refine biparts
1790 MB(11%)00:04:54 Iter 4 44.63% Refine biparts
1790 MB(11%)00:04:55 Iter 4 45.20% Refine biparts
1790 MB(11%)00:04:56 Iter 4 45.76% Refine biparts
1790 MB(11%)00:04:57 Iter 4 46.33% Refine biparts
1790 MB(11%)00:04:57 Iter 4 46.89% Refine biparts
1790 MB(11%)00:04:58 Iter 4 47.46% Refine biparts
1790 MB(11%)00:04:59 Iter 4 48.02% Refine biparts
1790 MB(11%)00:05:00 Iter 4 48.59% Refine biparts
1790 MB(11%)00:05:00 Iter 4 49.15% Refine biparts
1790 MB(11%)00:05:01 Iter 4 49.72% Refine biparts
1790 MB(11%)00:05:02 Iter 4 50.28% Refine biparts
1790 MB(11%)00:05:03 Iter 4 50.85% Refine biparts
1790 MB(11%)00:05:03 Iter 4 51.41% Refine biparts
1790 MB(11%)00:05:04 Iter 4 51.98% Refine biparts
1790 MB(11%)00:05:05 Iter 4 52.54% Refine biparts
1790 MB(11%)00:05:06 Iter 4 53.11% Refine biparts
1790 MB(11%)00:05:06 Iter 4 53.67% Refine biparts
1790 MB(11%)00:05:07 Iter 4 54.24% Refine biparts
1790 MB(11%)00:05:08 Iter 4 54.80% Refine biparts
1790 MB(11%)00:05:09 Iter 4 55.37% Refine biparts
1790 MB(11%)00:05:10 Iter 4 55.93% Refine biparts
1790 MB(11%)00:05:10 Iter 4 56.50% Refine biparts
1790 MB(11%)00:05:11 Iter 4 57.06% Refine biparts
1790 MB(11%)00:05:12 Iter 4 57.63% Refine biparts
1790 MB(11%)00:05:13 Iter 4 58.19% Refine biparts
1790 MB(11%)00:05:13 Iter 4 58.76% Refine biparts
1790 MB(11%)00:05:14 Iter 4 59.32% Refine biparts
1790 MB(11%)00:05:15 Iter 4 59.89% Refine biparts
1790 MB(11%)00:05:16 Iter 4 60.45% Refine biparts
1790 MB(11%)00:05:16 Iter 4 61.02% Refine biparts
1790 MB(11%)00:05:17 Iter 4 61.58% Refine biparts
1790 MB(11%)00:05:18 Iter 4 62.15% Refine biparts
1790 MB(11%)00:05:19 Iter 4 62.71% Refine biparts
1790 MB(11%)00:05:19 Iter 4 63.28% Refine biparts
1790 MB(11%)00:05:20 Iter 4 63.84% Refine biparts
1790 MB(11%)00:05:21 Iter 4 64.41% Refine biparts
1790 MB(11%)00:05:22 Iter 4 64.97% Refine biparts
1790 MB(11%)00:05:22 Iter 4 65.54% Refine biparts
1790 MB(11%)00:05:23 Iter 4 66.10% Refine biparts
1790 MB(11%)00:05:24 Iter 4 66.67% Refine biparts
1790 MB(11%)00:05:25 Iter 4 67.23% Refine biparts
1790 MB(11%)00:05:25 Iter 4 67.80% Refine biparts
1790 MB(11%)00:05:26 Iter 4 68.36% Refine biparts
1790 MB(11%)00:05:27 Iter 4 68.93% Refine biparts
1790 MB(11%)00:05:28 Iter 4 69.49% Refine biparts
1790 MB(11%)00:05:29 Iter 4 70.06% Refine biparts
1790 MB(11%)00:05:29 Iter 4 70.62% Refine biparts
1790 MB(11%)00:05:30 Iter 4 71.19% Refine biparts
1790 MB(11%)00:05:31 Iter 4 71.75% Refine biparts
1790 MB(11%)00:05:32 Iter 4 72.32% Refine biparts
1790 MB(11%)00:05:32 Iter 4 72.88% Refine biparts
1790 MB(11%)00:05:33 Iter 4 73.45% Refine biparts
1790 MB(11%)00:05:34 Iter 4 74.01% Refine biparts
1790 MB(11%)00:05:35 Iter 4 74.58% Refine biparts
1790 MB(11%)00:05:35 Iter 4 75.14% Refine biparts
1790 MB(11%)00:05:36 Iter 4 75.71% Refine biparts
1790 MB(11%)00:05:37 Iter 4 76.27% Refine biparts
1790 MB(11%)00:05:38 Iter 4 76.84% Refine biparts
1790 MB(11%)00:05:38 Iter 4 77.40% Refine biparts
1790 MB(11%)00:05:39 Iter 4 77.97% Refine biparts
1790 MB(11%)00:05:40 Iter 4 78.53% Refine biparts
1790 MB(11%)00:05:41 Iter 4 79.10% Refine biparts
1790 MB(11%)00:05:41 Iter 4 79.66% Refine biparts
1790 MB(11%)00:05:42 Iter 4 80.23% Refine biparts
1790 MB(11%)00:05:43 Iter 4 80.79% Refine biparts
1790 MB(11%)00:05:44 Iter 4 81.36% Refine biparts
1790 MB(11%)00:05:44 Iter 4 81.92% Refine biparts
1790 MB(11%)00:05:45 Iter 4 82.49% Refine biparts
1790 MB(11%)00:05:46 Iter 4 83.05% Refine biparts
1790 MB(11%)00:05:47 Iter 4 83.62% Refine biparts
1790 MB(11%)00:05:48 Iter 4 84.18% Refine biparts
1790 MB(11%)00:05:48 Iter 4 84.75% Refine biparts
1790 MB(11%)00:05:49 Iter 4 85.31% Refine biparts
1790 MB(11%)00:05:50 Iter 4 85.88% Refine biparts
1790 MB(11%)00:05:51 Iter 4 86.44% Refine biparts
1790 MB(11%)00:05:51 Iter 4 87.01% Refine biparts
1790 MB(11%)00:05:52 Iter 4 87.57% Refine biparts
1790 MB(11%)00:05:53 Iter 4 88.14% Refine biparts
1790 MB(11%)00:05:54 Iter 4 88.70% Refine biparts
1790 MB(11%)00:05:54 Iter 4 89.27% Refine biparts
1790 MB(11%)00:05:55 Iter 4 89.83% Refine biparts
1790 MB(11%)00:05:56 Iter 4 90.40% Refine biparts
1790 MB(11%)00:05:57 Iter 4 90.96% Refine biparts
1790 MB(11%)00:05:57 Iter 4 91.53% Refine biparts
1790 MB(11%)00:05:58 Iter 4 92.09% Refine biparts
1790 MB(11%)00:05:59 Iter 4 92.66% Refine biparts
1790 MB(11%)00:06:00 Iter 4 93.22% Refine biparts
1790 MB(11%)00:06:00 Iter 4 93.79% Refine biparts
1790 MB(11%)00:06:01 Iter 4 94.35% Refine biparts
1790 MB(11%)00:06:02 Iter 4 94.92% Refine biparts
1790 MB(11%)00:06:03 Iter 4 95.48% Refine biparts
1790 MB(11%)00:06:03 Iter 4 96.05% Refine biparts
1790 MB(11%)00:06:04 Iter 4 96.61% Refine biparts
1790 MB(11%)00:06:05 Iter 4 97.18% Refine biparts
1790 MB(11%)00:06:06 Iter 4 97.74% Refine biparts
1790 MB(11%)00:06:07 Iter 4 98.31% Refine biparts
1790 MB(11%)00:06:07 Iter 4 98.87% Refine biparts
1790 MB(11%)00:06:08 Iter 4 99.44% Refine biparts
1790 MB(11%)00:06:09 Iter 4 100.00% Refine biparts
1790 MB(11%)00:06:10 Iter 4 100.56% Refine biparts
1790 MB(11%)00:06:10 Iter 4 100.00% Refine bipartstipseq_aln <- DNAStringSet(tipseq_aln)## extract accession numbers from tip labels
tl <- tree$tip.label
acc <- sub("\\w+\\|", "", tl)
names(tl) <- acc
## writeXStringSet(tipseq_aln, file = "data/HPV58_aln.fas")
#tipseq_aln <- readDNAStringSet("data/HPV58_aln.fas")
tipseq_aln <- TDbook::dna_HPV58_aln %>%
as.character %>%
lapply(., paste0, collapse = "") %>%
unlist() %>%
Biostrings::DNAStringSet()## calculate pairwise hamming distances among sequences
tipseq_dist <- pwalign::stringDist(tipseq_aln, method = "hamming")
## calculate the percentage of differences
tipseq_d <- as.matrix(tipseq_dist) / width(tipseq_aln[1]) * 100
## convert the matrix to a tidy data frame for facet_plot
dd <- as_tibble(tipseq_d)
dd$seq1 <- rownames(tipseq_d)
td <- gather(dd,seq2, dist, -seq1)
td$seq1 <- tl[td$seq1]
td$seq2 <- tl[td$seq2]
g <- p$data$group
names(g) <- p$data$label
td$clade <- g[td$seq2]
## visualize the sequence differences using dot plot and line plot
## and align the sequence difference plot to the tree using facet_plot
p2 <- p + geom_facet(panel = "Sequence Distance",
data = td, geom = geom_point, alpha = .6,
mapping = aes(x = dist, color = clade, shape = clade)) +
geom_facet(panel = "Sequence Distance",
data = td, geom = geom_path, alpha = .6,
mapping=aes(x = dist, group = seq2, color = clade)) +
scale_shape_manual(values = 1:8, guide = FALSE)
print(p2)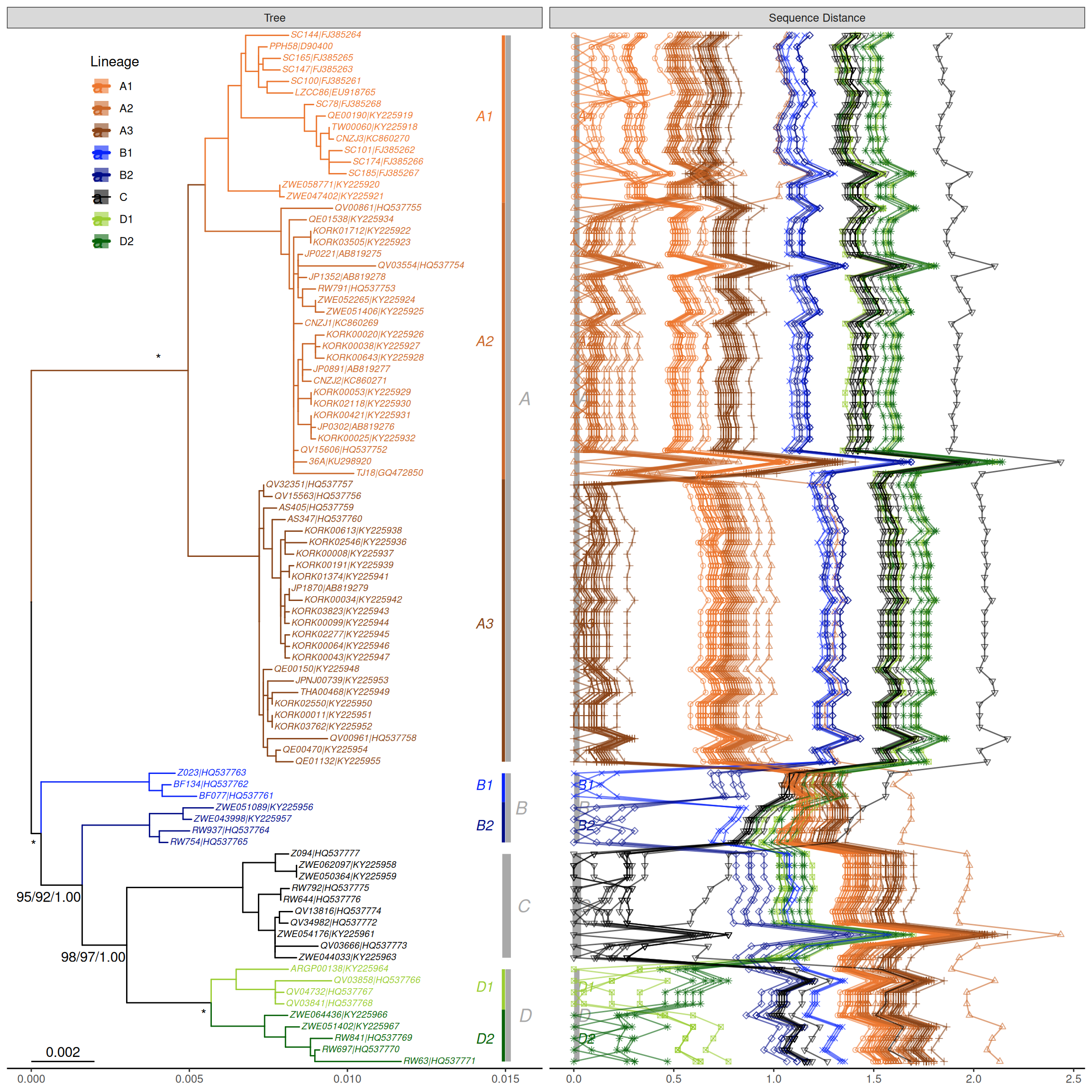
15.2 Displaying Different Symbolic Points for Bootstrap Values.
We can cut the bootstrap values into several intervals, e.g., to indicate whether the clade is of high, moderate, or low support. Then we can use these intervals as categorical variables to set different colors or shapes of symbolic points to indicate the bootstrap values belong to which category (Figure Figure 15.2).
## phytools also have a read.newick function
read.newick <- treeio::read.newicklibrary(treeio)
library(ggplot2)
library(ggtree)
library(TDbook)
tree <- read.newick(text=text_RMI_tree, node.label = "support")
root <- rootnode(tree)
ggtree(tree, color="black", size=1.5, linetype=1, right=TRUE) +
geom_tiplab(size=4.5, hjust = -0.060, fontface="bold") + xlim(0, 0.09) +
geom_point2(aes(subset=!isTip & node != root,
fill=cut(support, c(0, 700, 900, 1000))),
shape=21, size=4) +
theme_tree(legend.position=c(0.2, 0.2)) +
scale_fill_manual(values=c("white", "grey", "black"), guide='legend',
name='Bootstrap Percentage(BP)',
breaks=c('(900,1e+03]', '(700,900]', '(0,700]'),
labels=expression(BP>=90,70 <= BP * " < 90", BP < 70))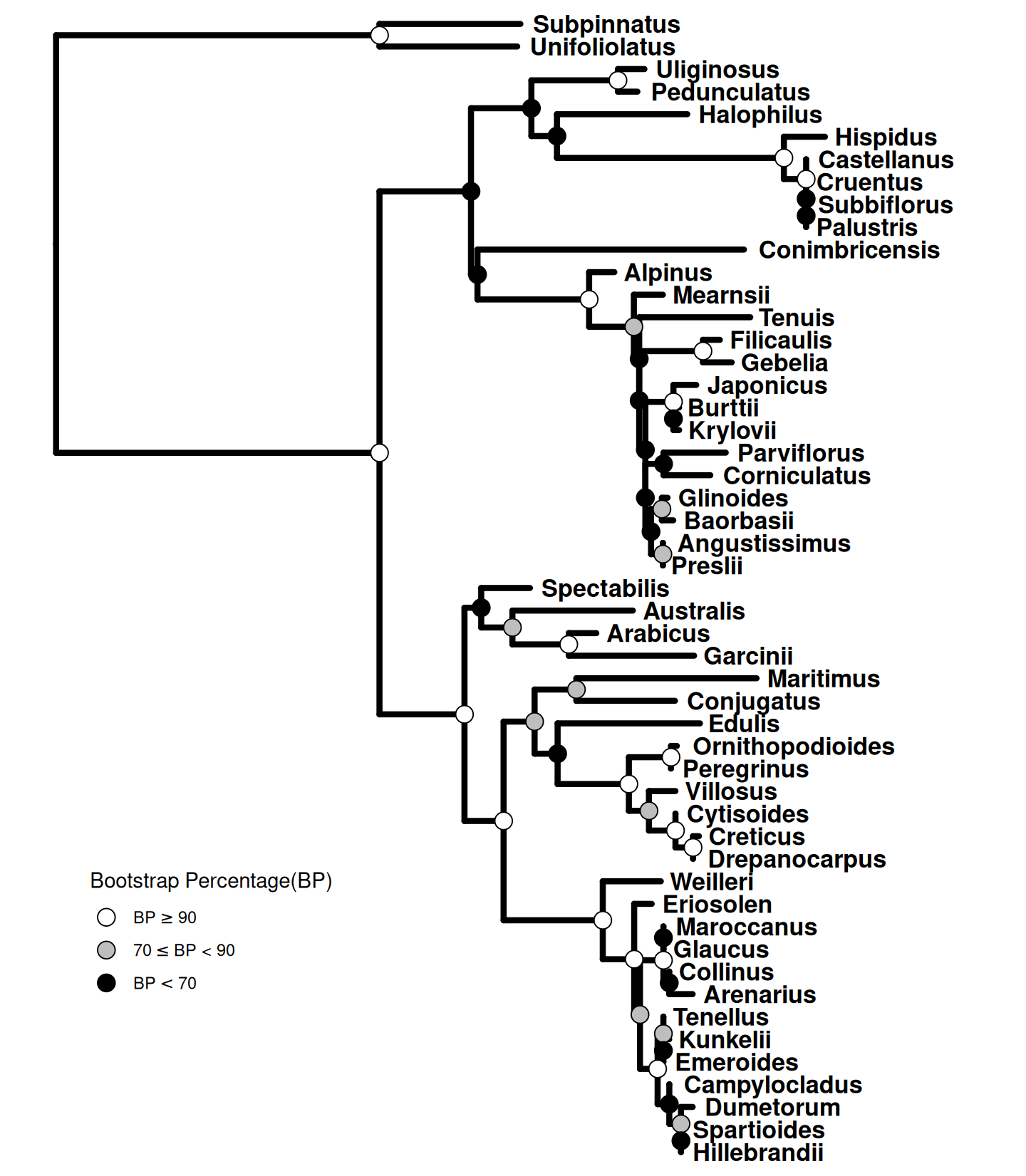
15.3 Highlighting Different Groups
This example reproduces Figure 1 of (Larsen et al., 2019). It used groupOTU() to add grouping information of chicken CTLDcps. The branch line type and color are defined based on this grouping information. Two groups of CTLDcps are highlighted in different background colors using geom_hilight (red for Group II and green for Group V). The avian-specific expansion of Group V with the subgroups of A and B- are labeled using geom_cladelab (Figure Figure 15.3).
library(TDbook)
mytree <- tree_treenwk_30.4.19
# Define nodes for coloring later on
tiplab <- mytree$tip.label
cls <- tiplab[grep("^ch", tiplab)]
labeltree <- groupOTU(mytree, cls)
p <- ggtree(labeltree, aes(color=group, linetype=group), layout="circular") +
scale_color_manual(values = c("#efad29", "#63bbd4")) +
geom_nodepoint(color="black", size=0.1) +
geom_tiplab(size=2, color="black")
p2 <- flip(p, 136, 110) %>%
flip(141, 145) %>%
rotate(141) %>%
rotate(142) %>%
rotate(160) %>%
rotate(164) %>%
rotate(131)
### Group V and II coloring
dat <- data.frame(
node = c(110, 88, 156,136),
fill = c("#229f8a", "#229f8a", "#229f8a", "#f9311f")
)
p3 <- p2 +
geom_hilight(
data = dat,
mapping = aes(
node = node,
fill = I(fill)
),
alpha = 0.2,
extendto = 1.4
)
### Putting on a label on the avian specific expansion
p4 <- p3 +
geom_cladelab(
node = 113,
label = "Avian-specific expansion",
align = TRUE,
angle = -35,
offset.text = 0.05,
hjust = "center",
fontsize = 2,
offset = .2,
barsize = .2
)
### Adding the bootstrap values with subset used to remove all bootstraps < 50
p5 <- p4 +
geom_nodelab(
mapping = aes(
x = branch,
label = label,
subset = !is.na(as.numeric(label)) & as.numeric(label) > 50
),
size = 2,
color = "black",
nudge_y = 0.6
)
### Putting labels on the subgroups
p6 <- p5 +
geom_cladelab(
data = data.frame(
node = c(114, 121),
name = c("Subgroup A", "Subgroup B")
),
mapping = aes(
node = node,
label = name
),
align = TRUE,
offset = .05,
offset.text = .03,
hjust = "center",
barsize = .2,
fontsize = 2,
angle = "auto",
horizontal = FALSE
) +
theme(
legend.position = "none",
plot.margin = grid::unit(c(-15, -15, -15, -15), "mm")
)
print(p6)Warning in FUN(X[[i]], ...): NAs introduced by coercion
Warning in FUN(X[[i]], ...): NAs introduced by coercion
Warning in FUN(X[[i]], ...): NAs introduced by coercion
Warning in FUN(X[[i]], ...): NAs introduced by coercion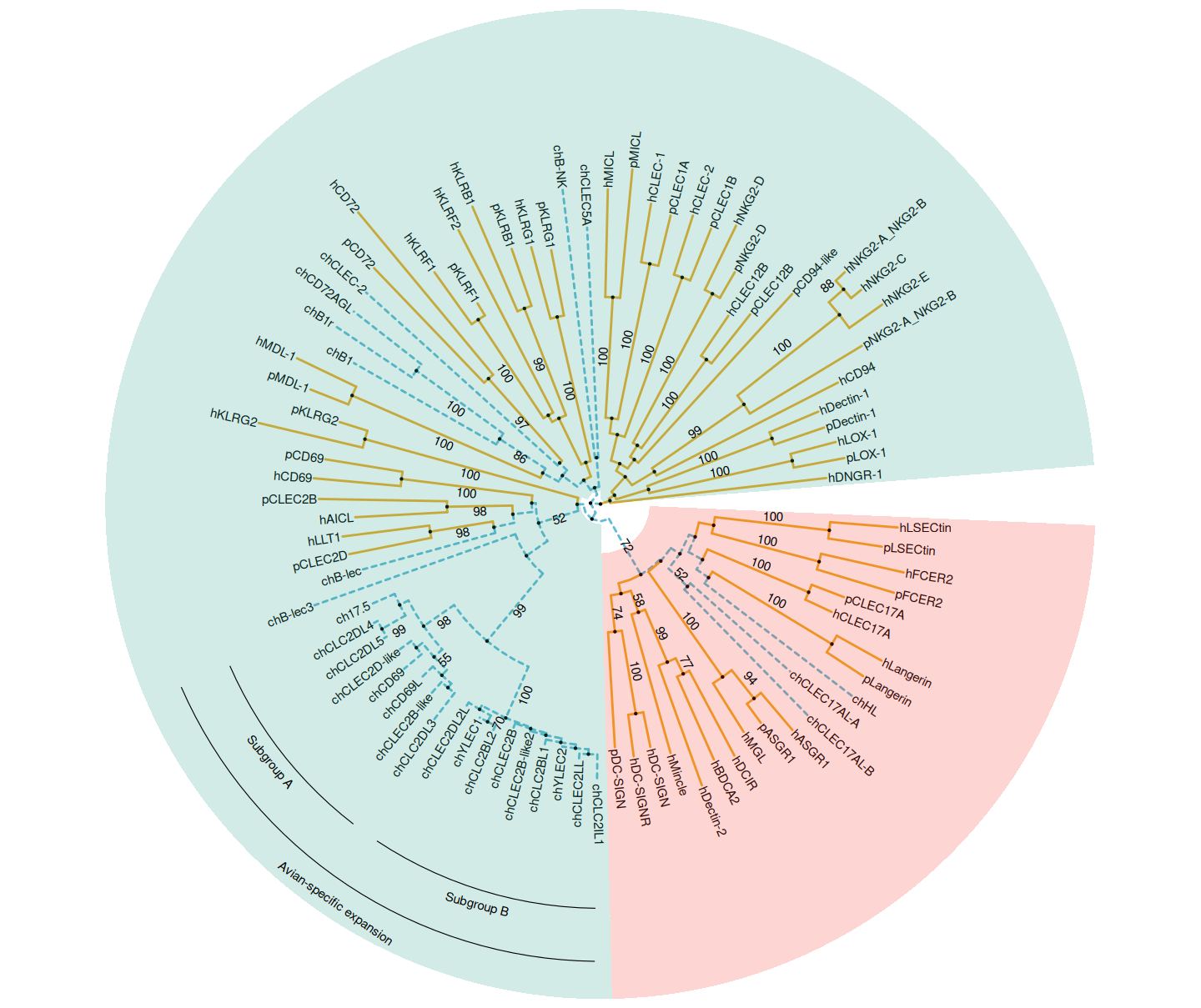
15.4 Phylogenetic Tree with Genome Locus Structure
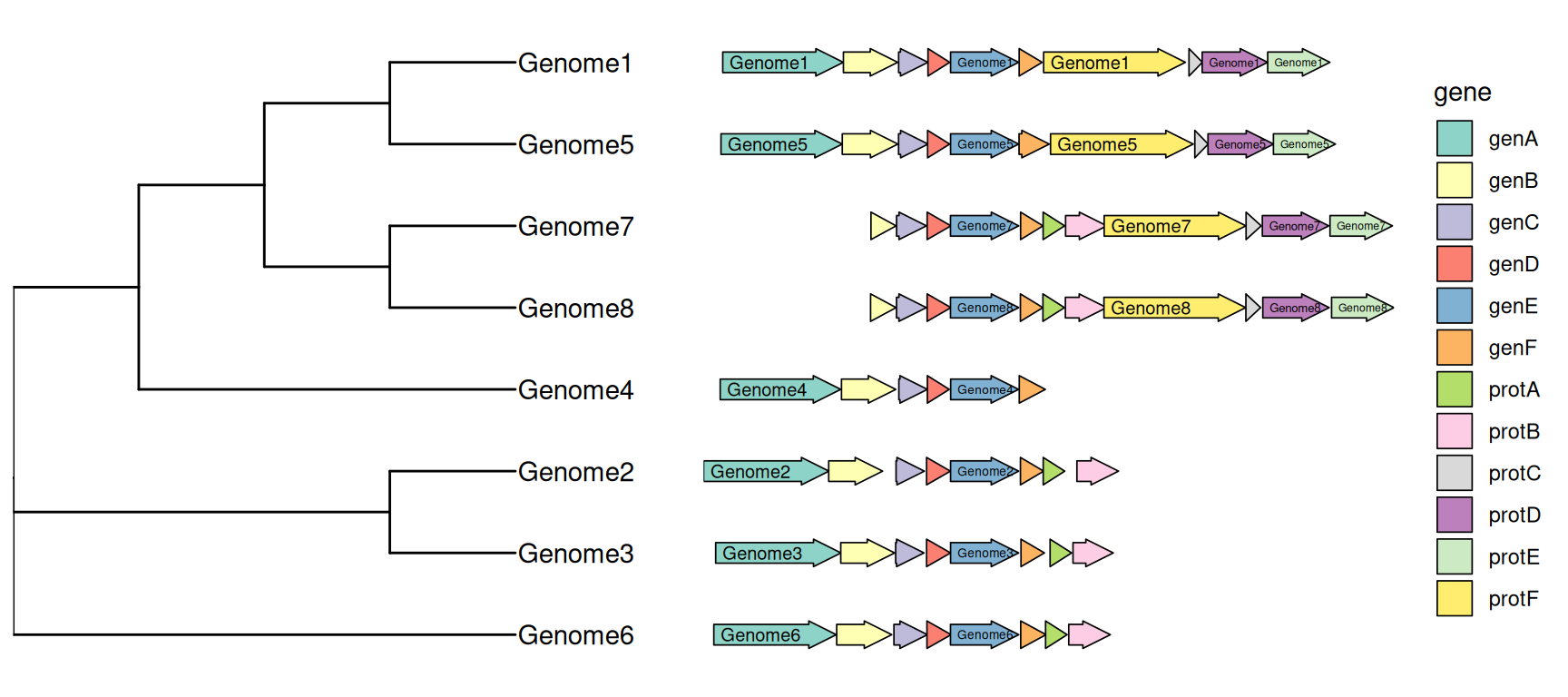
The geom_motif() is defined in ggtree and it is a wrapper layer of the gggenes::geom_gene_arrow(). The geom_motif() can automatically adjust genomic alignment by selective gene (via the on parameter) and can label genes via the label parameter. In the following example, we use example_genes dataset provided by gggenes. As the dataset only provides genomic coordination of a set of genes, a phylogeny for the genomes needs to be constructed first. We calculate Jaccard similarity based on the ratio of overlapping genes among genomes and correspondingly determine genome distance. The BioNJ algorithm was applied to construct the tree. Then we can use geom_facet() to visualize the tree with the genomic structures (Figure Figure 15.4).
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:Biostrings':
collapse, intersect, setdiff, setequal, unionThe following object is masked from 'package:GenomeInfoDb':
intersectThe following object is masked from 'package:XVector':
sliceThe following objects are masked from 'package:IRanges':
collapse, desc, intersect, setdiff, slice, unionThe following objects are masked from 'package:S4Vectors':
first, intersect, rename, setdiff, setequal, unionThe following objects are masked from 'package:BiocGenerics':
combine, intersect, setdiff, unionThe following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(ggplot2)
library(gggenes)
library(ggtree)
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
for (i in 1:n) {
for (j in 1:i) {
jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
length(union(genes[[i]], genes[[j]]))
d[j, i] <- d[i, j] <- 1 - jaccard_sim
}
}
tree <- ape::bionj(d)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))Warning in x[i] <- value: number of items to replace is not a multiple of
replacement length