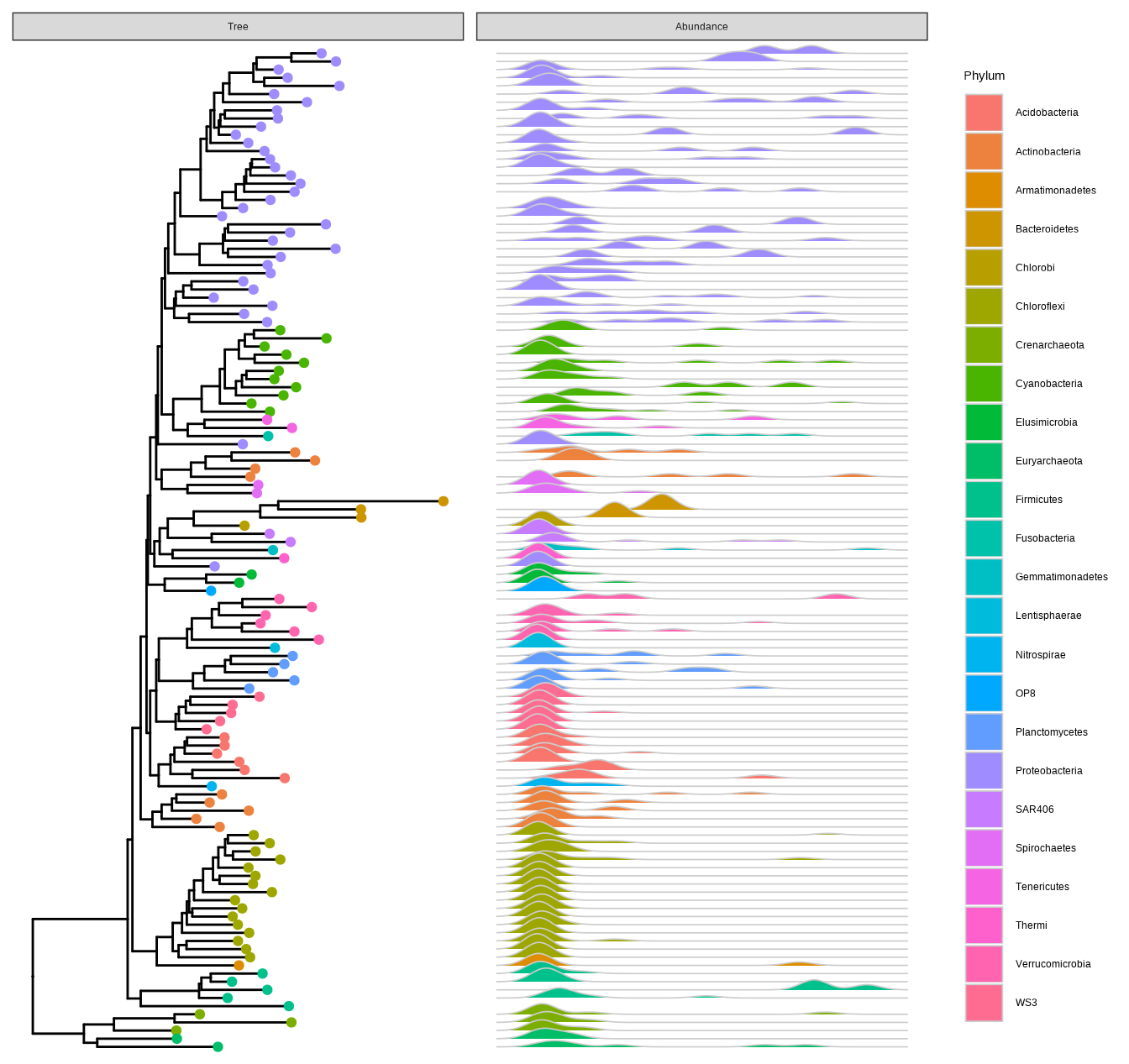
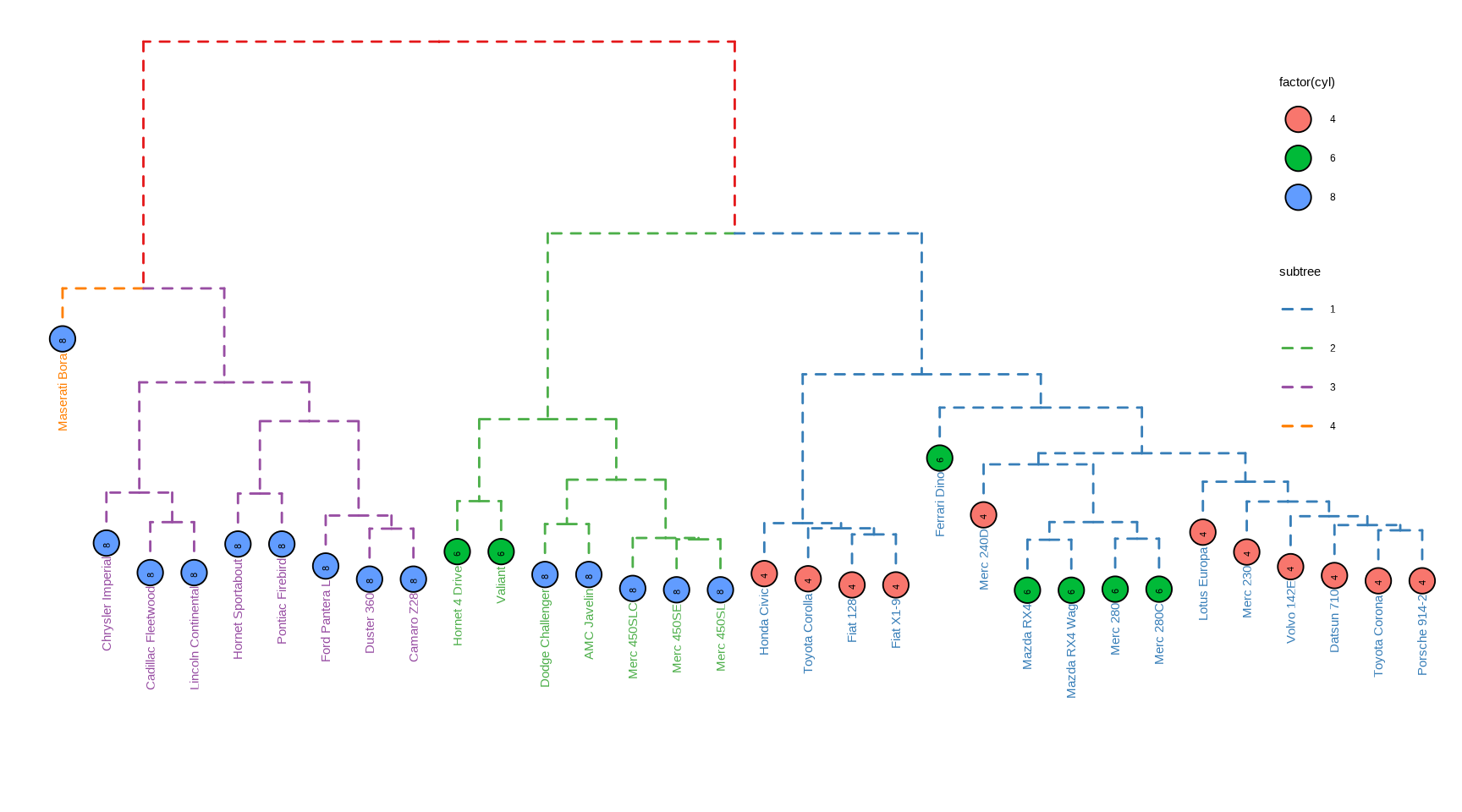
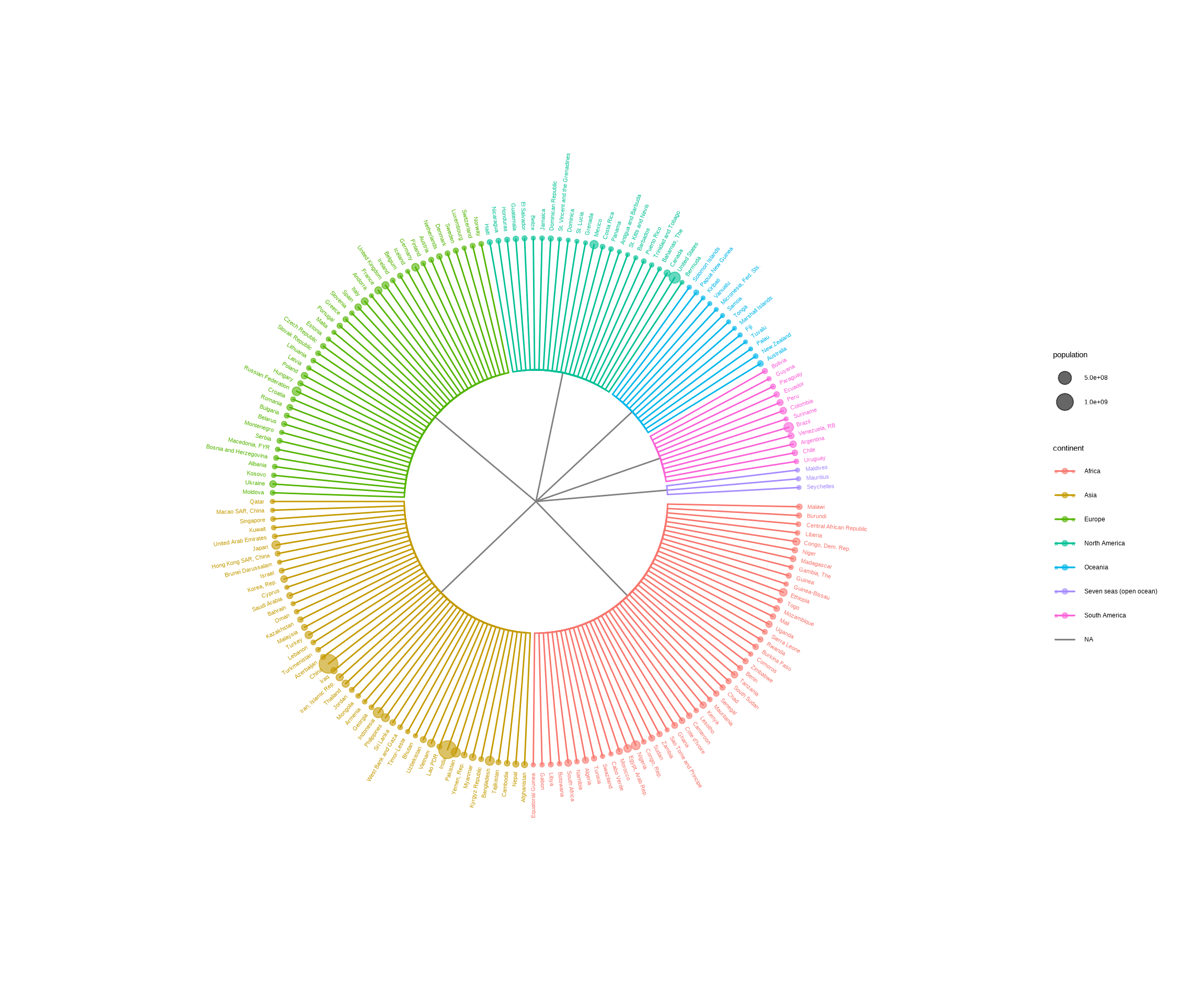
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., & Holmes, S. P. (2016).
DADA2:
High-resolution sample inference from
Illumina amplicon data.
Nature Methods,
13(7), 581–583.
https://doi.org/10.1038/nmeth.3869
Kuczynski, J., Stombaugh, J., Walters, W. A., González, A., Caporaso, J. G., & Knight, R. (2011). Using
QIIME to analyze 16S
rRNA gene sequences from
Microbial Communities.
Current Protocols in Bioinformatics / Editoral Board, Andreas D. Baxevanis ... [Et Al.],
CHAPTER, Unit10.7.
https://doi.org/10.1002/0471250953.bi1007s36
Kunin, V., & Hugenholtz, P. (2010).
PyroTagger :
A fast , accurate pipeline for analysis of
rRNA amplicon pyrosequence data.
The Open Journal, 1–8.
http://www.theopenjournal.org/toj_articles/1
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., & Weber, C. F. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities.
Applied and Environmental Microbiology,
75(23), 7537–7541.
https://doi.org/10.1128/AEM.01541-09
Wang, L.-G., Lam, T. T.-Y., Xu, S., Dai, Z., Zhou, L., Feng, T., Guo, P., Dunn, C. W., Jones, B. R., Bradley, T., Zhu, H., Guan, Y., Jiang, Y., & Yu, G. (2020). Treeio: An r package for phylogenetic tree input and output with richly annotated and associated data.
Molecular Biology and Evolution,
37(2), 599–603.
https://doi.org/10.1093/molbev/msz240
Yu, G. (2020). Using ggtree to visualize data on tree-like structures.
Current Protocols in Bioinformatics,
69(1), e96.
https://doi.org/10.1002/cpbi.96
Yu, G., Lam, T. T.-Y., Zhu, H., & Guan, Y. (2018). Two methods for mapping and visualizing associated data on phylogeny using ggtree.
Molecular Biology and Evolution,
35(12), 3041–3043.
https://doi.org/10.1093/molbev/msy194
Yu, G., Smith, D. K., Zhu, H., Guan, Y., & Lam, T. T.-Y. (2017). Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data.
Methods in Ecology and Evolution,
8(1), 28–36.
https://doi.org/10.1111/2041-210X.12628