3 Coarse-graining of large single-cell data into metacells using SuperCell
Here, we use the pbmc4k dataset as presented in Batch correction.
3.1 RunSuperCell
The RunSuperCell() function is a wrapper function to run SuperCell.
## class: SingleCellExperiment
## dim: 33694 842
## metadata(3): hvgmethod hvgcols SuperCell
## assays(2): counts logcounts
## rownames(33694): ENSG00000243485 ENSG00000237613 ...
## ENSG00000277475 ENSG00000268674
## rowData names(7): ENSEMBL_ID Symbol_TENx ...
## variance.expected variance.standardized
## colnames: NULL
## colData names(1): size
## reducedDimNames(0):
## mainExpName: NULL
## altExpNames(0):The output of RunSuperCell() is a SingleCellExperiment object that stores gene expression matrix of metacells.
We can use it for downstram analysis.
post_process <- function(pbmc) {
pbmc <- NormalizeData(pbmc)
pbmc <- FindVariableFeatures(pbmc)
pbmc <- ScaleData(pbmc)
pbmc <- runPCA(pbmc, subset_row = VariableFeatures(pbmc), exprs_values = "scaled")
pbmc <- FindNeighbors(pbmc, dims = 1:10)
pbmc <- FindClusters(pbmc)
pbmc <- RunUMAP(pbmc)
return(pbmc)
}
pbmc2 <- post_process(pbmc)
library(ggsc)
sc_dim(pbmc2, reduction="UMAP") + sc_dim_geom_label()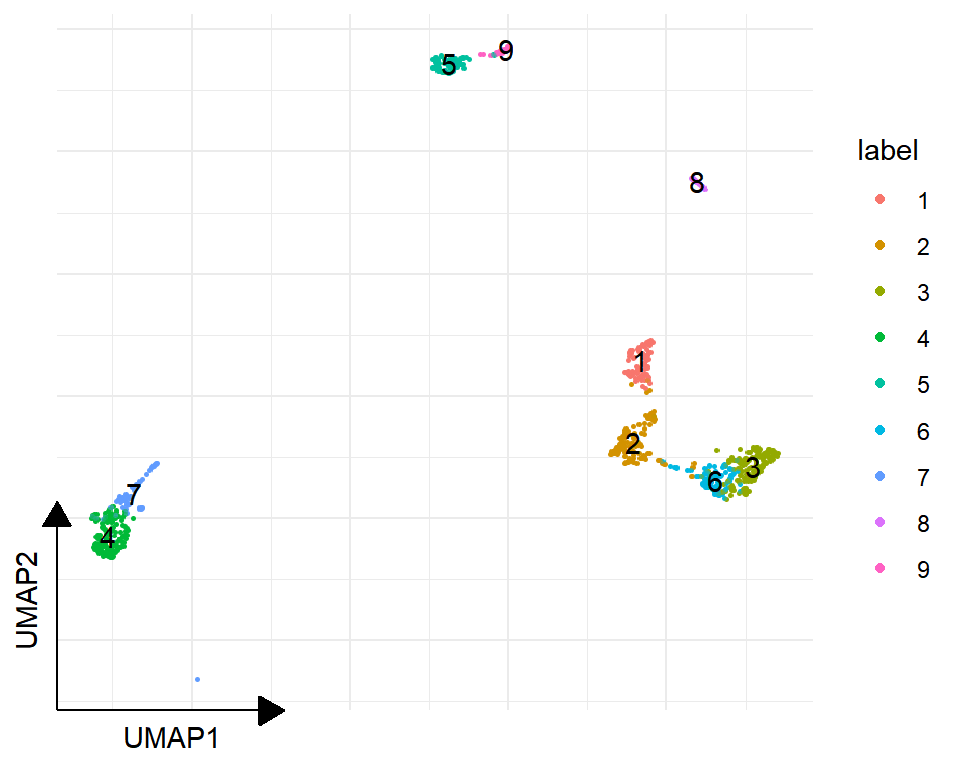
3.2 Estimate SuperCell purity
pbmc4k2 <- post_process(pbmc4k)
SC <- metadata(pbmc2)$SuperCell
purity <- SuperCell::supercell_purity(pbmc4k2$label, SC$membership, method = 'entropy')
head(purity)## 1 2 3 4 5 6
## 0.0000000 1.0397208 0.4505612 1.0549202 0.3046361 0.5004024## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.0000 0.0000 0.0000 0.1805 0.4506 1.3863