1 Basic: preprocessing and clustering
We created Seurat-like functions to help you get started effortlessly.
1.2 QC
# colData(pbmc)$percent.mt <- PercentageFeatureSet(pbmc, "^MT-")
pbmc[["percent.mt"]] <- PercentageFeatureSet(pbmc, "^MT-")
VlnPlot(pbmc, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"))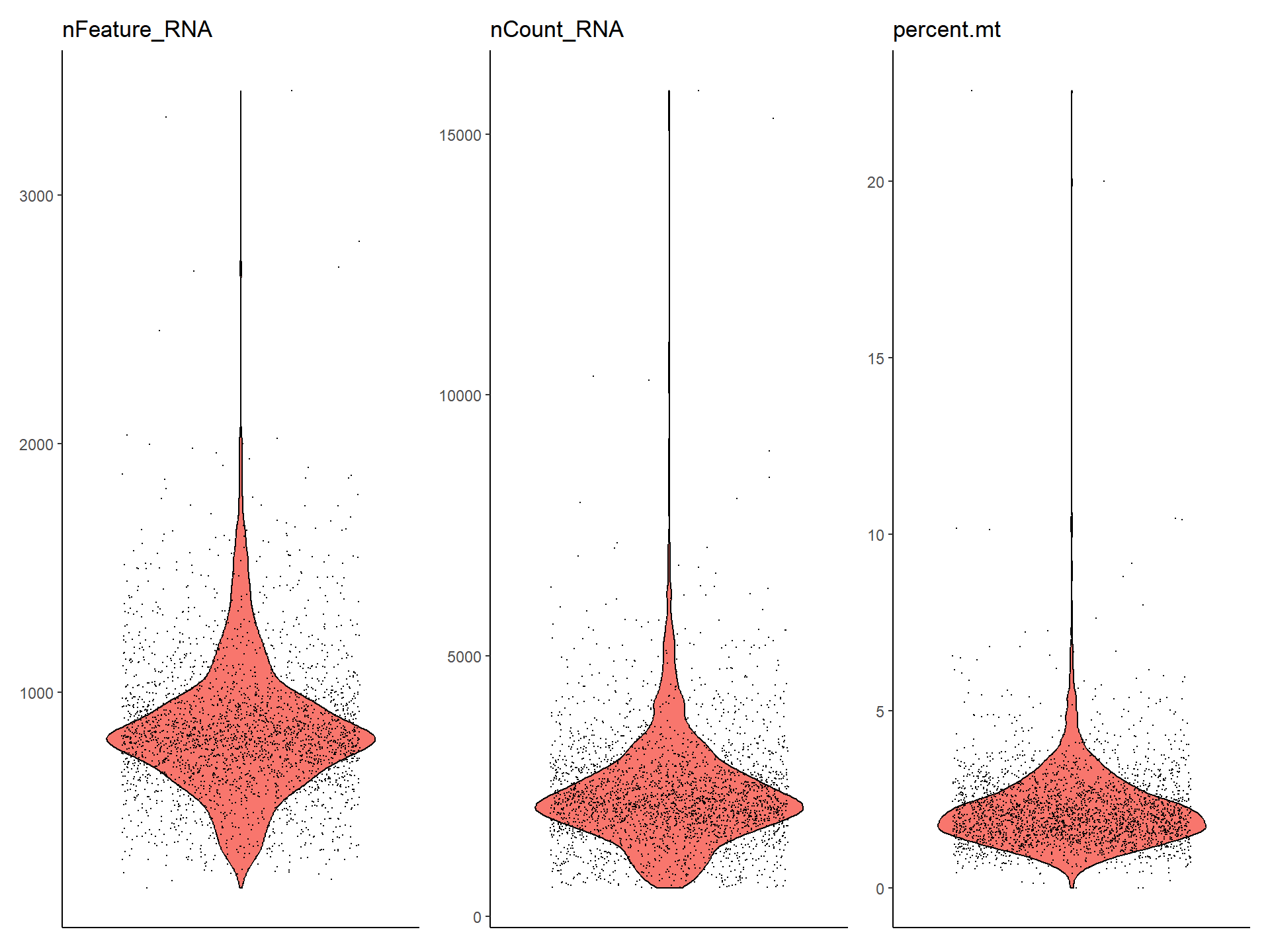
## ************************************************** >=0 | 32738 (100%)
## ************************* >=1 | 16634 (51%)
## ********************** >=2 | 14702 (45%)
## ********************* >=3 | 13714 (42%)
## ******************** >=4 | 13034 (40%)
## ******************* >=5 | 12572 (38%)
## ******************* >=6 | 12198 (37%)
## ****************** >=7 | 11876 (36%)
## ****************** >=8 | 11627 (36%)
## ***************** >=9 | 11379 (35%)
## ***************** >=10 | 11139 (34%)pbmc <- subset_feature(pbmc, mincell = 3, peek=FALSE)
pbmc <- subset(pbmc, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5)
pbmc## class: SingleCellExperiment
## dim: 13714 2638
## metadata(1): Samples
## assays(1): counts
## rownames(13714): AL627309.1 AP006222.2 ...
## PNRC2_ENSG00000215700 SRSF10_ENSG00000215699
## rowData names(2): ID Symbol
## colnames: NULL
## colData names(5): Sample Barcode nFeature_RNA
## nCount_RNA percent.mt
## reducedDimNames(0):
## mainExpName: NULL
## altExpNames(0):plot1 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "percent.mt")
plot2 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "nFeature_RNA")
library(aplot)
plot_list(plot1, plot2)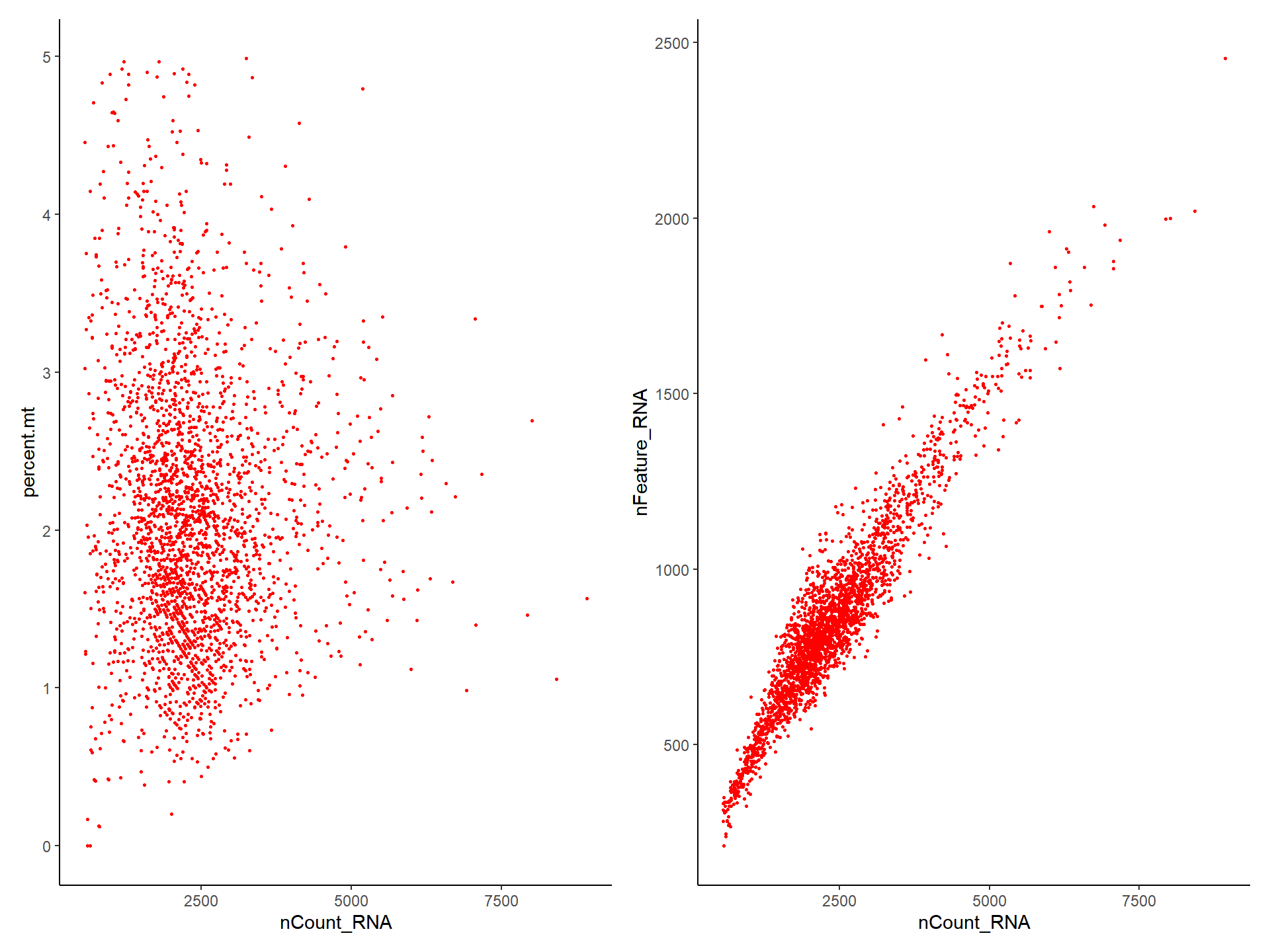
Users can also use plotColData() to visualize the QC data.
# VlnPlot-like
p1 <- plotColData(pbmc, y = 'nCount_RNA')
# FeatureScatter-like
p2 <- plotColData(pbmc, x = "nCount_RNA", y = "percent.mt")
plot_list(p1, p2)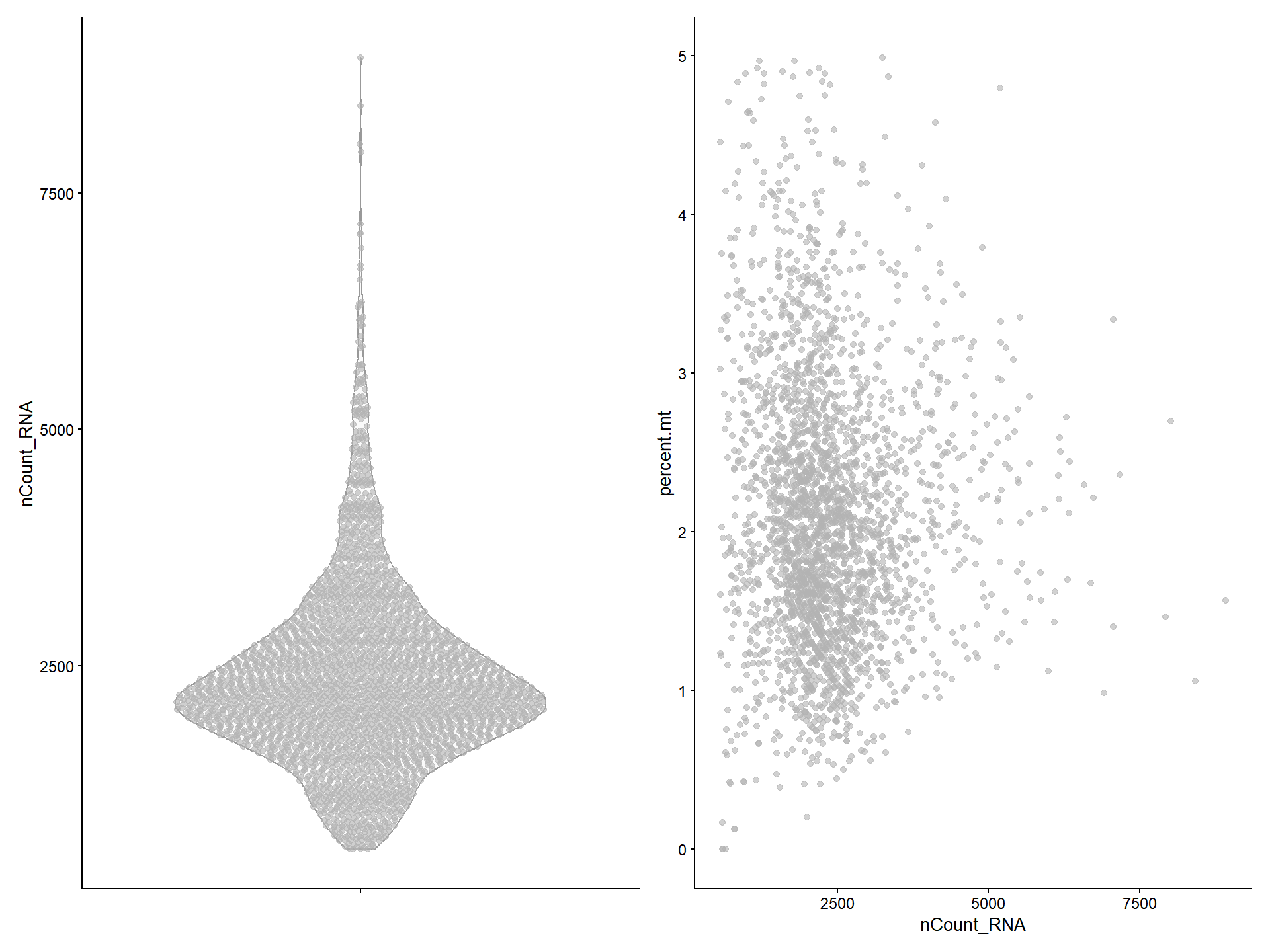
1.3 Variable features
pbmc <- NormalizeData(pbmc)
pbmc <- FindVariableFeatures(pbmc, method = "seurat")
top10 <- head(VariableFeatures(pbmc, method = "seurat"), 10)
top10## [1] "PPBP" "LYZ" "S100A9" "IGLL5" "GNLY" "FTL"
## [7] "PF4" "FTH1" "GNG11" "S100A8"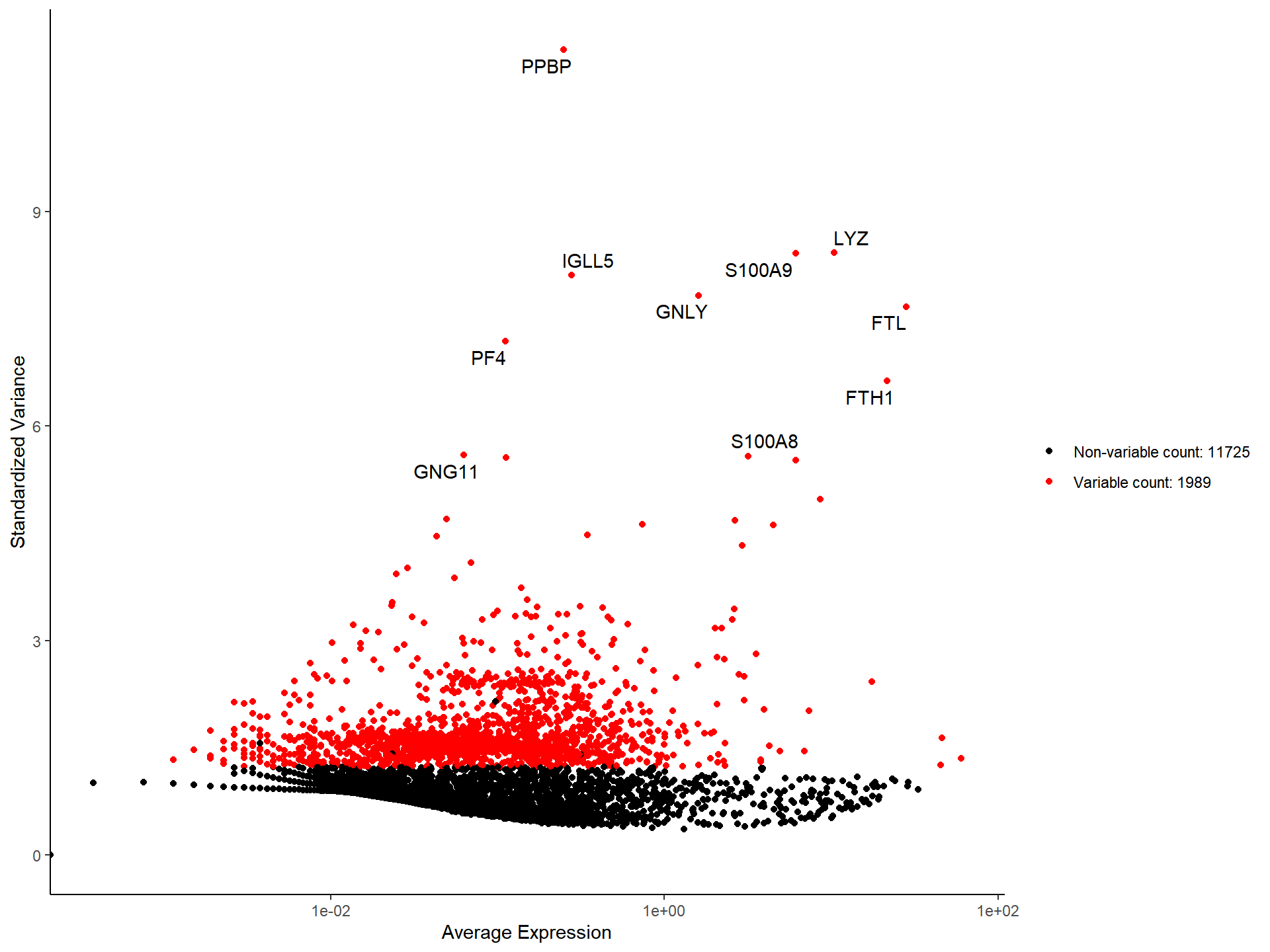
1.4 Dimensional reduction
pbmc <- ScaleData(pbmc)
pbmc <- runPCA(pbmc, subset_row = VariableFeatures(pbmc, method = "seurat"), exprs_values = "scaled")
ElbowPlot(pbmc)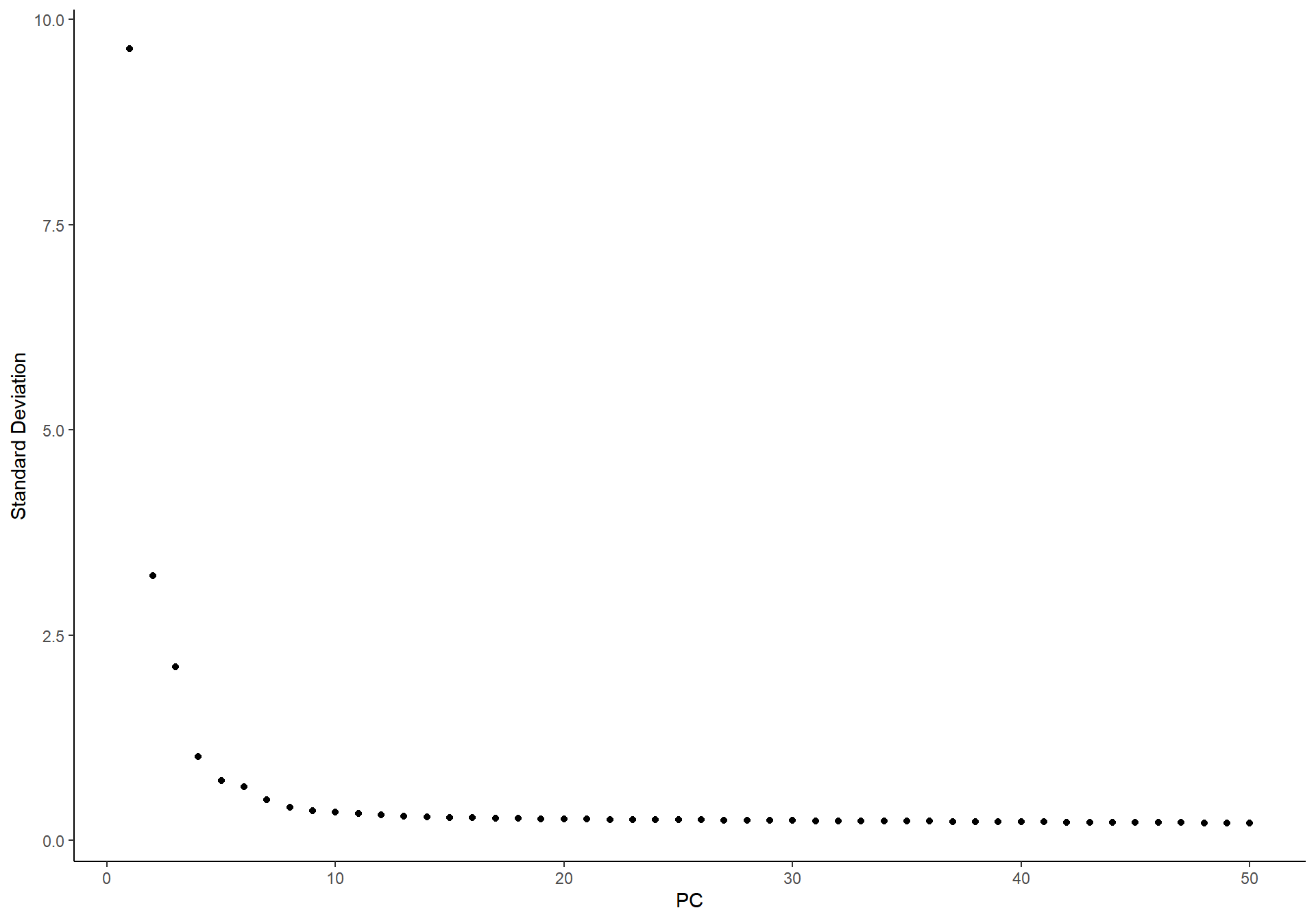
1.5 Clustering
set.seed(2024-10-01)
pbmc <- FindNeighbors(pbmc, dims = 1:10)
pbmc <- FindClusters(pbmc, resolution = 0.88)
head(Idents(pbmc), 5)## [1] 1 2 3 4 5
## Levels: 1 2 3 4 5 6 7 8 91.6 UMAP
pbmc <- RunUMAP(pbmc, 1:10)
library(aplot)
# DimPlot is a wrapper for ggsc::sc_dim
p1 <- DimPlot(pbmc, reduction="PCA")
p2 <- DimPlot(pbmc, reduction="UMAP")
plot_list(gglist = list(PCA=p1, UMAP=p2))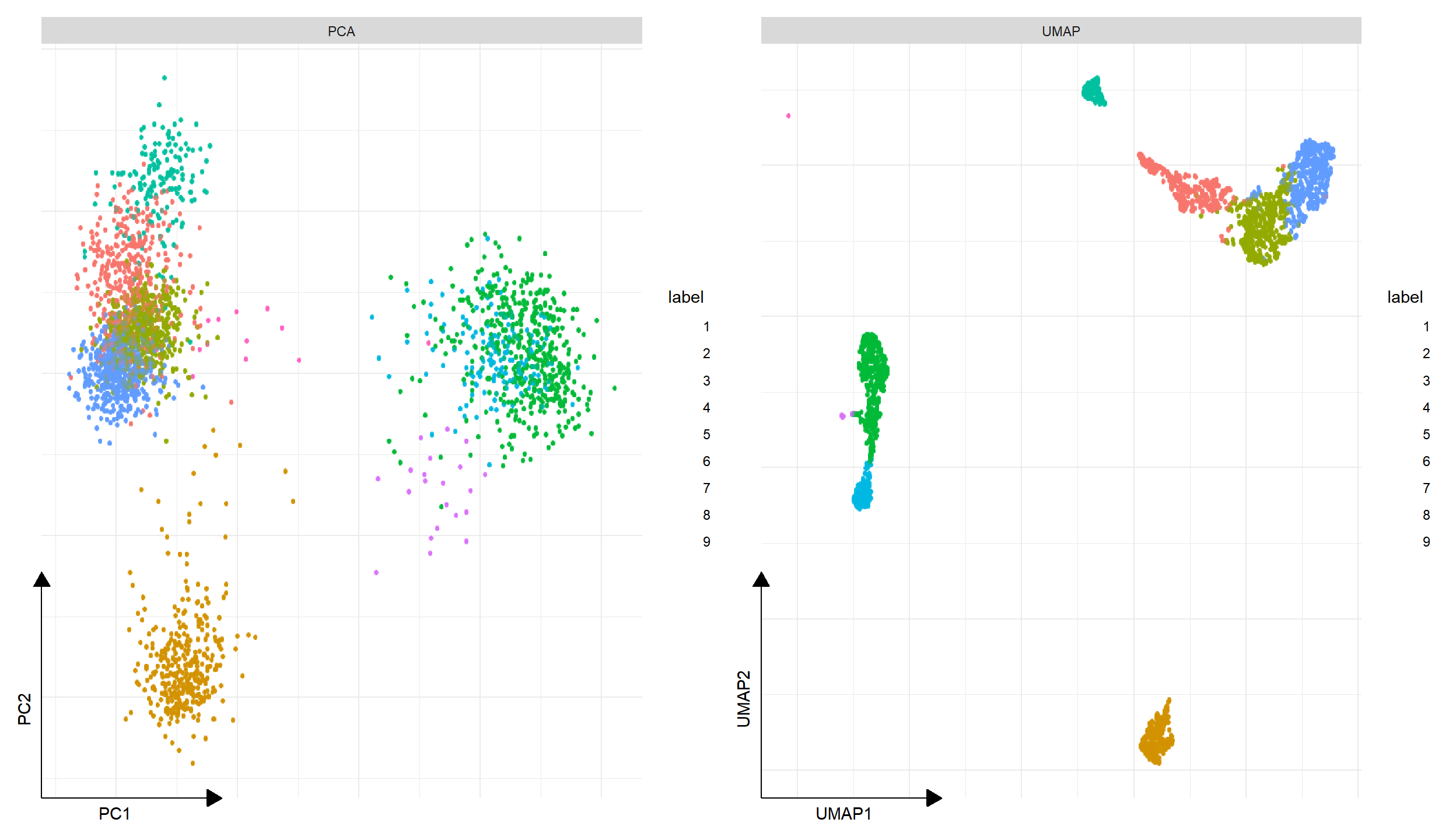
1.7 Find Markers
# find all markers of cluster 2
cluster2.markers <- FindMarkers(pbmc, ident.1 = 2)
head(cluster2.markers, n = 5)## pval padj avg_log2FC pct.1
## CD79A 0.000000e+00 0.000000e+00 7.984223 0.936
## MS4A1 0.000000e+00 0.000000e+00 6.403330 0.855
## CD79B 2.655974e-274 1.214134e-270 5.391680 0.916
## LINC00926 2.397625e-272 8.220258e-269 8.120877 0.564
## TCL1A 9.481783e-271 2.600664e-267 7.427626 0.622
## pct.2
## CD79A 0.041
## MS4A1 0.053
## CD79B 0.142
## LINC00926 0.009
## TCL1A 0.022# find all markers distinguishing cluster 5 from clusters 0 and 3
cluster5.markers <- FindMarkers(pbmc, ident.1 = 5, ident.2 = c(0, 3))
head(cluster5.markers, n = 5)## pval padj avg_log2FC pct.1 pct.2
## NKG7 1.936869e-125 2.656222e-121 7.031062 0.933 0.160
## CCL5 2.338439e-114 1.603468e-110 4.775635 0.949 0.243
## CST7 1.442905e-91 6.596000e-88 4.832158 0.759 0.105
## GZMA 1.066563e-89 3.656713e-86 3.989925 0.759 0.098
## GZMK 1.850701e-73 5.076103e-70 4.284187 0.640 0.076# find markers for every cluster compared to all remaining cells,
# report only the positive ones
library(dplyr)
pbmc.markers <- FindAllMarkers(pbmc, only.pos = TRUE)## Calculating markers for all clusters using presto...topN_marker <- function(markers, n) {
markers %>%
group_by(cluster) %>%
dplyr::filter(avg_log2FC > 1) %>%
slice_head(n = n) %>%
ungroup()
}
top10 <- topN_marker(pbmc.markers, 10)
split(top10$gene, top10$cluster)## $`1`
## [1] "LDHB" "CD3D" "CD3E" "LTB"
##
## $`2`
## [1] "CD79A" "MS4A1" "CD79B" "LINC00926"
## [5] "TCL1A" "HLA-DQA1" "VPREB3" "HLA-DQB1"
## [9] "CD74" "HLA-DRA"
##
## $`3`
## [1] "LDHB" "LTB" "IL32" "IL7R" "CD3D" "CD3E"
## [7] "CD2" "JUNB" "TMEM66" "GIMAP7"
##
## $`4`
## [1] "LGALS2" "MS4A6A" "CD14" "GRN" "FCN1" "CST3"
## [7] "ALDH2" "CFP" "CPVL" "TYMP"
##
## $`5`
## [1] "GZMK" "CCL5" "NKG7" "CST7" "GZMA" "CTSW" "CD8A"
## [8] "LYAR" "IL32" "KLRG1"
##
## $`6`
## [1] "MALAT1"
##
## $`7`
## [1] "CDKN1C" "HES4" "RP11-290F20.3"
## [4] "MS4A7" "FCGR3A" "LILRA3"
## [7] "IFITM3" "PILRA" "LRRC25"
## [10] "LILRB1"
##
## $`8`
## [1] "S100A8" "S100A9" "LGALS2" "TYROBP" "FCN1" "FTL"
## [7] "CST3" "LYZ" "GPX1" "RBP7"
##
## $`9`
## [1] "GZMB" "FGFBP2" "PRF1" "SPON2" "CST7" "GZMA"
## [7] "GZMH" "GNLY" "CCL4" "AKR1C3"1.7.1 Marker gene information
top1 <- topN_marker(pbmc.markers, 1)
gene_table <- gene_summary_table(top1, gene_col = 'gene',
cluster_col = 'cluster', keyType='SYMBOL', output = "data.frame")## Please cite:
##
## D Wang, G Chen, L Li, S Wen, Z Xie, X Luo, L
## Zhan, S Xu, J Li, R Wang, Q Wang, G Yu. Reducing language
## barriers, promoting information absorption, and
## communication using fanyi. Chinese Medical Journal. 2024,
## 137(16):1950-1956. doi: 10.1097/CM9.0000000000003242## ## 'select()' returned 1:1 mapping between keys and
## columns## ENTREZID gene pval padj avg_log2FC
## 1 1028 CDKN1C 1.689064e-212 2.316382e-208 1.136389
## 2 3002 GZMB 0.000000e+00 0.000000e+00 4.770946
## 3 3003 GZMK 3.460655e-181 4.745943e-177 2.398711
## 4 378938 MALAT1 4.208396e-26 5.771394e-22 1.040261
## 5 3945 LDHB 4.964439e-69 4.004842e-66 1.456801
## 6 3945 LDHB 5.609128e-109 7.692358e-105 1.748761
## 7 3957 LGALS2 6.272118e-221 8.601582e-217 3.246368
## 8 6279 S100A8 1.938236e-194 2.658097e-190 5.241459
## 9 973 CD79A 0.000000e+00 0.000000e+00 3.930314
## pct.1 pct.2 cluster avgExpr statistic auc name
## 1 0.506 0.010 7 1.160143 299976.5 0.7478622 CDKN1C
## 2 0.968 0.056 9 4.934502 447991.0 0.9774271 GZMB
## 3 0.640 0.057 5 2.574675 483425.0 0.8011617 GZMK
## 4 1.000 1.000 6 8.828117 209041.0 0.8081315 MALAT1
## 5 0.917 0.622 1 3.501892 769929.0 0.7530958 LDHB
## 6 0.966 0.591 3 3.662064 961320.0 0.7936712 LDHB
## 7 0.952 0.130 4 3.709701 589423.5 0.9218961 LGALS2
## 8 0.949 0.204 8 6.016170 576490.5 0.9453888 S100A8
## 9 0.936 0.041 2 4.027329 761864.5 0.9654413 CD79A
## description
## 1 cyclin dependent kinase inhibitor 1C
## 2 granzyme B
## 3 granzyme K
## 4 metastasis associated lung adenocarcinoma transcript 1
## 5 lactate dehydrogenase B
## 6 lactate dehydrogenase B
## 7 galectin 2
## 8 S100 calcium binding protein A8
## 9 CD79a molecule
## summary
## 1 This gene is imprinted, with preferential expression of the maternal allele. The encoded protein is a tight-binding, strong inhibitor of several G1 cyclin/Cdk complexes and a negative regulator of cell proliferation. Mutations in this gene are implicated in sporadic cancers and Beckwith-Wiedemann syndorome, suggesting that this gene is a tumor suppressor candidate. Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Oct 2010]
## 2 This gene encodes a member of the granzyme subfamily of proteins, part of the peptidase S1 family of serine proteases. The encoded preproprotein is secreted by natural killer (NK) cells and cytotoxic T lymphocytes (CTLs) and proteolytically processed to generate the active protease, which induces target cell apoptosis. This protein also processes cytokines and degrades extracellular matrix proteins, and these roles are implicated in chronic inflammation and wound healing. Expression of this gene may be elevated in human patients with cardiac fibrosis. [provided by RefSeq, Sep 2016]
## 3 This gene product is a member of a group of related serine proteases from the cytoplasmic granules of cytotoxic lymphocytes. Cytolytic T lymphocytes (CTL) and natural killer (NK) cells share the remarkable ability to recognize, bind, and lyse specific target cells. They are thought to protect their host by lysing cells bearing on their surface 'nonself' antigens, usually peptides or proteins resulting from infection by intracellular pathogens. The protein described here lacks consensus sequences for N-glycosylation present in other granzymes. [provided by RefSeq, Jul 2008]
## 4 This gene produces a precursor transcript from which a long non-coding RNA is derived by RNase P cleavage of a tRNA-like small ncRNA (known as mascRNA) from its 3' end. The resultant mature transcript lacks a canonical poly(A) tail but is instead stabilized by a 3' triple helical structure. This transcript is retained in the nucleus where it is thought to form molecular scaffolds for ribonucleoprotein complexes. It may act as a transcriptional regulator for numerous genes, including some genes involved in cancer metastasis and cell migration, and it is involved in cell cycle regulation. Its upregulation in multiple cancerous tissues has been associated with the proliferation and metastasis of tumor cells. [provided by RefSeq, Mar 2015]
## 5 This gene encodes the B subunit of lactate dehydrogenase enzyme, which catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of NADH and NAD+ in a post-glycolysis process. Alternatively spliced transcript variants have been found for this gene. Recent studies have shown that a C-terminally extended isoform is produced by use of an alternative in-frame translation termination codon via a stop codon readthrough mechanism, and that this isoform is localized in the peroxisomes. Mutations in this gene are associated with lactate dehydrogenase B deficiency. Pseudogenes have been identified on chromosomes X, 5 and 13. [provided by RefSeq, Feb 2016]
## 6 This gene encodes the B subunit of lactate dehydrogenase enzyme, which catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of NADH and NAD+ in a post-glycolysis process. Alternatively spliced transcript variants have been found for this gene. Recent studies have shown that a C-terminally extended isoform is produced by use of an alternative in-frame translation termination codon via a stop codon readthrough mechanism, and that this isoform is localized in the peroxisomes. Mutations in this gene are associated with lactate dehydrogenase B deficiency. Pseudogenes have been identified on chromosomes X, 5 and 13. [provided by RefSeq, Feb 2016]
## 7 The protein encoded by this gene is a soluble beta-galactoside binding lectin. The encoded protein is found as a homodimer and can bind to lymphotoxin-alpha. A single nucleotide polymorphism in an intron of this gene can alter the transcriptional level of the protein, with a resultant increased risk of myocardial infarction. [provided by RefSeq, Jul 2008]
## 8 The protein encoded by this gene is a member of the S100 family of proteins containing 2 EF-hand calcium-binding motifs. S100 proteins are localized in the cytoplasm and/or nucleus of a wide range of cells, and involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation. S100 genes include at least 13 members which are located as a cluster on chromosome 1q21. This protein may function in the inhibition of casein kinase and as a cytokine. Altered expression of this protein is associated with the disease cystic fibrosis. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2016]
## 9 The B lymphocyte antigen receptor is a multimeric complex that includes the antigen-specific component, surface immunoglobulin (Ig). Surface Ig non-covalently associates with two other proteins, Ig-alpha and Ig-beta, which are necessary for expression and function of the B-cell antigen receptor. This gene encodes the Ig-alpha protein of the B-cell antigen component. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]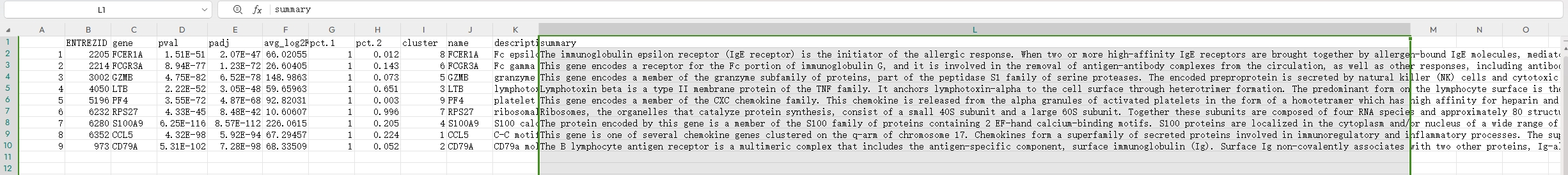
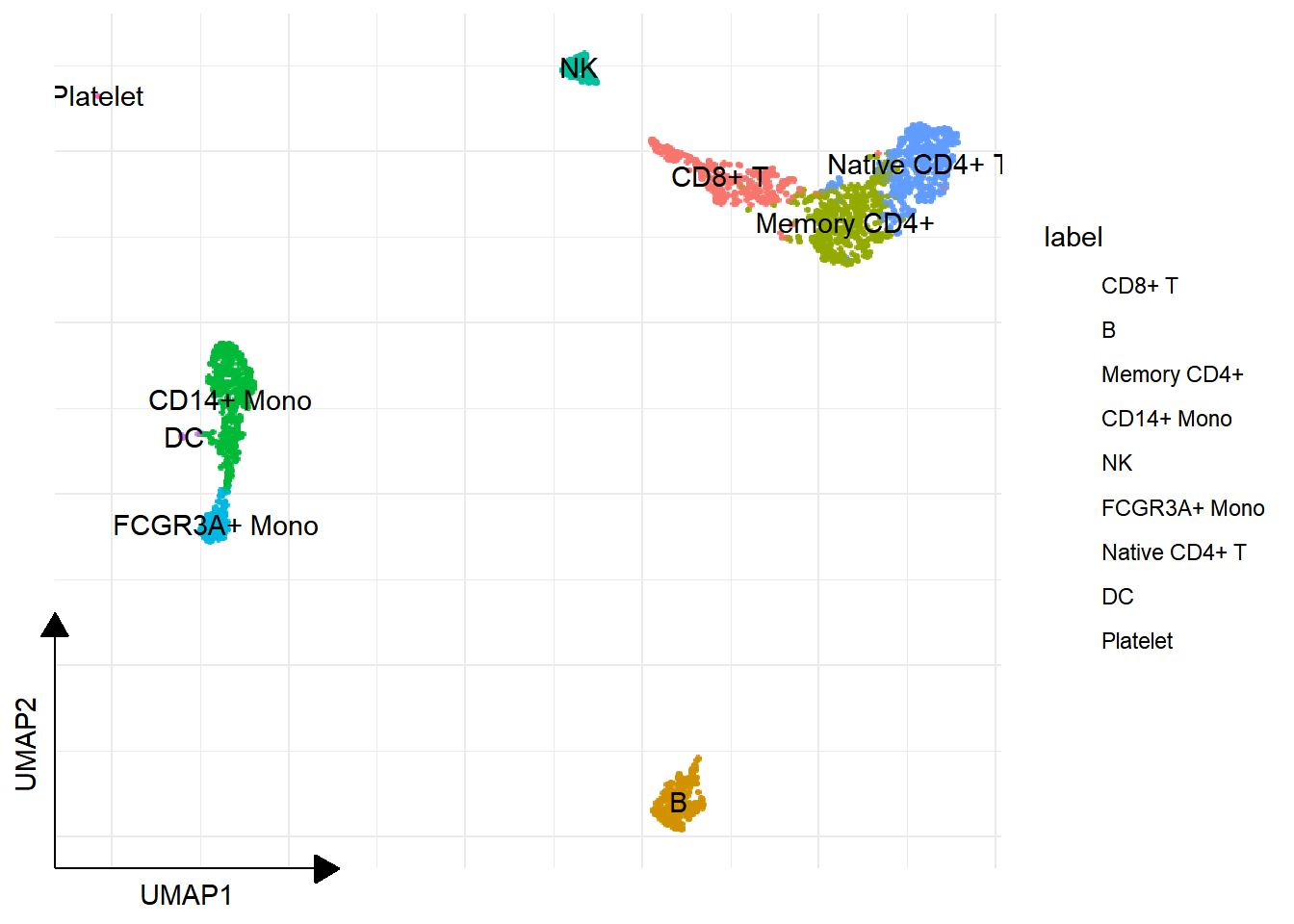
1.8 Cell cluster annotation
new_labels <- c(
"1" = "Naive CD4+ T",
"2" = "B",
"3" = "Memory CD4+",
"4" = "CD14+ Mono",
"5" = "NK",
"6" = "CD8+ T",
"7" = "Stress Response / Malignant Cells",
"8" = "FCGR3A+ Mono",
"9" = "Myeloid / Neutrophil"
)
pbmc <- RenameIdents(pbmc, new_labels)
DimPlot(pbmc, reduction="UMAP") + ggsc::sc_dim_geom_label()