4 Cell type annotation
4.1 Manual annotation
Please refer to the Cell cluster annotation session.
4.2 Automatic annotation
4.2.1 Run SingleR
## class: SummarizedExperiment
## dim: 19363 713
## metadata(0):
## assays(1): logcounts
## rownames(19363): A1BG A1BG-AS1 ... ZZEF1 ZZZ3
## rowData names(0):
## colnames(713): GSM112490 GSM112491 ... GSM92233
## GSM92234
## colData names(3): label.main label.fine label.ont## DataFrame with 6 rows and 4 columns
## scores labels delta.next
## <matrix> <character> <numeric>
## 1 0.1040907:0.233575:0.222844:... T_cells 0.0888618
## 2 0.1017499:0.358524:0.299235:... B_cell 0.3046119
## 3 0.0696533:0.265561:0.237793:... T_cells 0.1985708
## 4 0.0863639:0.235842:0.275857:... Monocyte 0.0503585
## 5 0.0737555:0.164465:0.175442:... NK_cell 0.0892196
## 6 0.1281878:0.258801:0.249245:... T_cells 0.0775251
## pruned.labels
## <character>
## 1 T_cells
## 2 B_cell
## 3 T_cells
## 4 Monocyte
## 5 NK_cell
## 6 T_cellsdice <- celldex::DatabaseImmuneCellExpressionData()
res2 <- SingleR(test = assay(pbmc, "logcounts"), ref = dice, labels = dice$label.main)
head(res2)## DataFrame with 6 rows and 4 columns
## scores labels
## <matrix> <character>
## 1 0.01278759:0.0594162:0.1731098:... T cells, CD4+
## 2 0.26468267:0.1424061:0.0667195:... B cells
## 3 0.03184648:0.0768767:0.2039776:... T cells, CD4+
## 4 -0.00914745:0.3439882:0.0610696:... Monocytes
## 5 -0.04285154:0.0396558:0.2513840:... NK cells
## 6 0.06123675:0.0980547:0.1470082:... T cells, CD4+
## delta.next pruned.labels
## <numeric> <character>
## 1 0.00808206 T cells, CD4+
## 2 0.12227658 B cells
## 3 0.05825145 T cells, CD4+
## 4 0.28291862 Monocytes
## 5 0.15577995 NK cells
## 6 0.12673642 T cells, CD4+4.2.3 Comparision with manual annotation result
# Ensure manual labels are available in colData for comparison
pbmc$label <- colLabels(pbmc)
x <- colData(pbmc)[, c("label", "hpca_label", "dice_label")]
plot_list(
manual = sc_dim(pbmc, reduction="UMAP"),
hpca = sc_dim(pbmc, reduction="UMAP", mapping=aes(color=hpca_label)),
dice = sc_dim(pbmc, reduction="UMAP", mapping=aes(color=dice_label)),
ncol = 3) &
sc_dim_geom_label(geom = ggrepel::geom_text_repel) &
theme(legend.position='inside')## Found more than one class "package_version" in cache; using the first, from namespace 'SeuratObject'## Also defined by 'alabaster.base'## Found more than one class "package_version" in cache; using the first, from namespace 'SeuratObject'## Also defined by 'alabaster.base'## Found more than one class "package_version" in cache; using the first, from namespace 'SeuratObject'## Also defined by 'alabaster.base'## Found more than one class "package_version" in cache; using the first, from namespace 'SeuratObject'## Also defined by 'alabaster.base'## Found more than one class "package_version" in cache; using the first, from namespace 'SeuratObject'## Also defined by 'alabaster.base'## Found more than one class "package_version" in cache; using the first, from namespace 'SeuratObject'## Also defined by 'alabaster.base'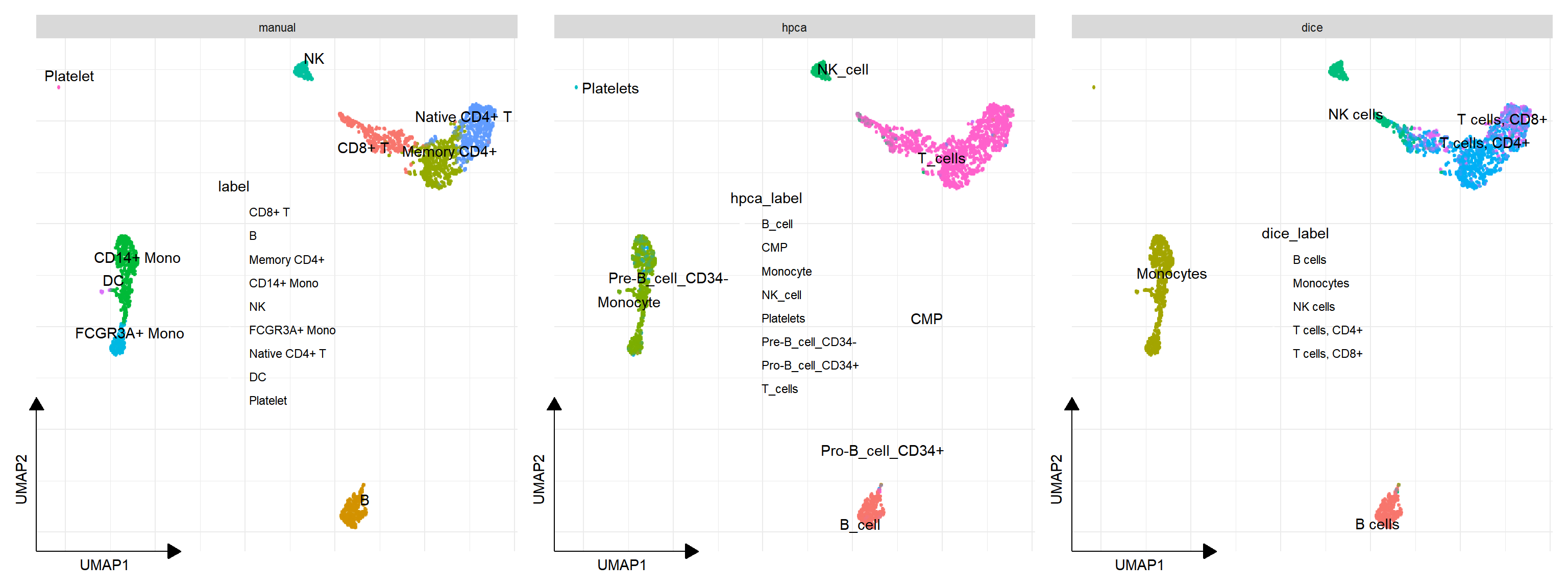
## hpca_label
## label B_cell CMP Monocyte
## Naive CD4+ T 0 0 0
## B 333 0 0
## Memory CD4+ 0 4 0
## CD14+ Mono 0 0 252
## NK 0 0 0
## CD8+ T 0 0 0
## Stress Response / Malignant Cells 0 0 155
## FCGR3A+ Mono 0 0 208
## Myeloid / Neutrophil 0 0 0
## hpca_label
## label NK_cell Platelets
## Naive CD4+ T 1 0
## B 0 0
## Memory CD4+ 1 0
## CD14+ Mono 0 0
## NK 29 0
## CD8+ T 4 0
## Stress Response / Malignant Cells 0 0
## FCGR3A+ Mono 0 12
## Myeloid / Neutrophil 154 0
## hpca_label
## label Pre-B_cell_CD34-
## Naive CD4+ T 1
## B 1
## Memory CD4+ 2
## CD14+ Mono 17
## NK 1
## CD8+ T 2
## Stress Response / Malignant Cells 6
## FCGR3A+ Mono 35
## Myeloid / Neutrophil 0
## hpca_label
## label Pro-B_cell_CD34+
## Naive CD4+ T 0
## B 4
## Memory CD4+ 0
## CD14+ Mono 0
## NK 0
## CD8+ T 0
## Stress Response / Malignant Cells 0
## FCGR3A+ Mono 1
## Myeloid / Neutrophil 0
## hpca_label
## label T_cells
## Naive CD4+ T 470
## B 6
## Memory CD4+ 585
## CD14+ Mono 1
## NK 223
## CD8+ T 96
## Stress Response / Malignant Cells 1
## FCGR3A+ Mono 0
## Myeloid / Neutrophil 33## dice_label
## label B cells Monocytes
## Naive CD4+ T 0 0
## B 340 0
## Memory CD4+ 0 4
## CD14+ Mono 0 269
## NK 0 0
## CD8+ T 0 0
## Stress Response / Malignant Cells 0 162
## FCGR3A+ Mono 0 256
## Myeloid / Neutrophil 0 0
## dice_label
## label NK cells T cells, CD4+
## Naive CD4+ T 2 273
## B 1 2
## Memory CD4+ 3 505
## CD14+ Mono 0 1
## NK 128 97
## CD8+ T 12 46
## Stress Response / Malignant Cells 0 0
## FCGR3A+ Mono 0 0
## Myeloid / Neutrophil 186 1
## dice_label
## label T cells, CD8+
## Naive CD4+ T 197
## B 1
## Memory CD4+ 80
## CD14+ Mono 0
## NK 28
## CD8+ T 44
## Stress Response / Malignant Cells 0
## FCGR3A+ Mono 0
## Myeloid / Neutrophil 0