2 Batch Correction
2.2 Preprocess
library(sclet)
sce_process <- function(sce) {
sce <- QCMetrics(sce)
sce[["percent.mt"]] <- PercentageFeatureSet(sce, "^MT-")
sce <- subset(sce, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5)
sce <- NormalizeData(sce)
sce <- FindVariableFeatures(sce)
return(sce)
}
pbmc3k <- sce_process(pbmc3k)
pbmc4k <- sce_process(pbmc4k)
pbmc_list <- list(pbmc3k = pbmc3k, pbmc4k = pbmc4k)2.4 Clustering
sce_clustering <- function(sce, assay = "logcounts", dims = 1:10) {
sce <- scater::runPCA(sce, subset_row = VariableFeatures(sce), exprs_values = assay)
sce <- FindNeighbors(sce, dims = dims)
sce <- FindClusters(sce)
sce <- RunUMAP(sce, dims = dims)
return(sce)
}
combined_pbmc <- sce_clustering(combined_pbmc, assay="logcounts")
rm_batch_pbmc <- sce_clustering(rm_batch_pbmc, assay = "reconstructed")2.5 Visualization
library(ggplot2)
library(ggsc)
library(aplot)
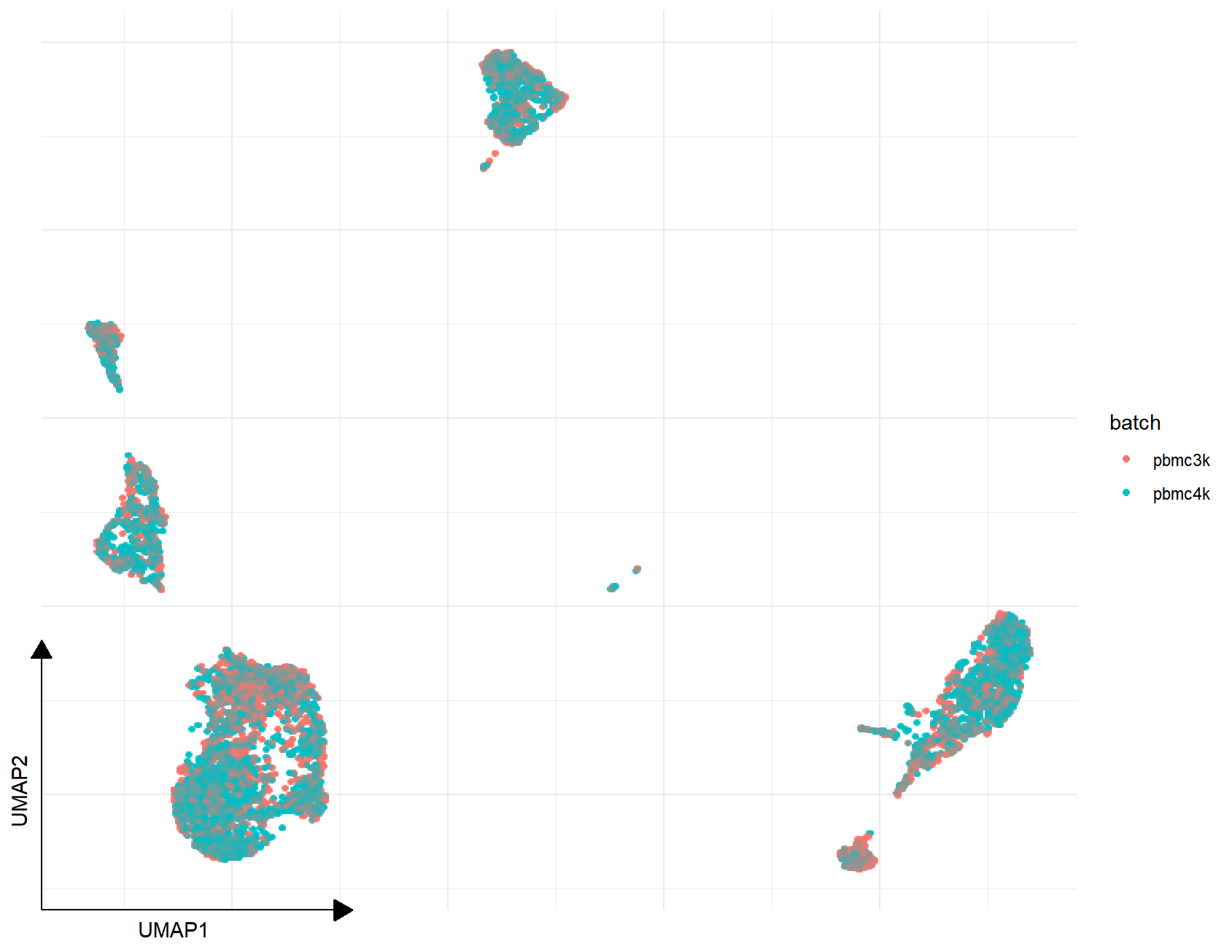
p1 <- sc_dim(combined_pbmc, reduction="UMAP", mapping=aes(color=batch))
p2 <- sc_dim(rm_batch_pbmc, reduction="UMAP", mapping=aes(color=batch))
p3 <- plot_list(Before=p1, After=p2)
p4 <- sc_dim(rm_batch_pbmc, reduction="UMAP")
plot_list(p3, p4, ncol=1, heights=c(1, 2))