ggmsa is designed for visualization and annotation of multiple sequence alignment. It implements functions to visualize publication-quality multiple sequence alignments (protein/DNA/RNA) in R extremely simple and powerful.
For details, please visit http://yulab-smu.top/ggmsa/
🔨 Installation
~~You can install ggmsa from CRAN using install.packages('ggmsa'). ~~
Alternatively you can grab the development version from github using devtools:
if (!requireNamespace("devtools", quietly=TRUE))
install.packages("devtools")
devtools::install_github("YuLab-SMU/ggmsa")💡 Quick Example
library(ggmsa)
protein_sequences <- system.file("extdata", "sample.fasta", package = "ggmsa")
ggmsa(protein_sequences, start = 221, end = 280, char_width = 0.5, seq_name = T) + geom_seqlogo() + geom_msaBar()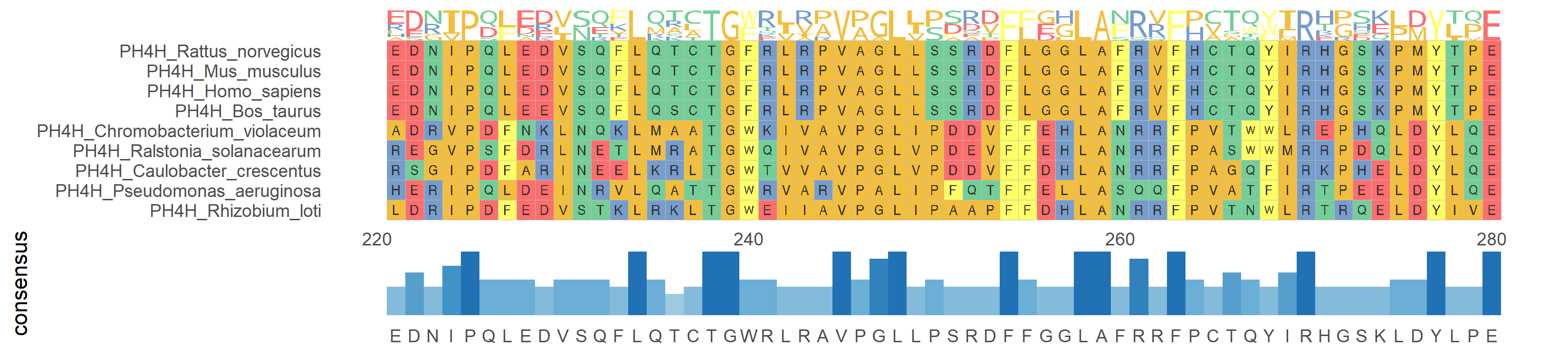
📚 Learn more
Check out the guides for learning everything there is to know about all the different features:
💖 Contributing
We welcome any contributions! By participating in this project you agree to abide by the terms outlined in the Contributor Code of Conduct.



