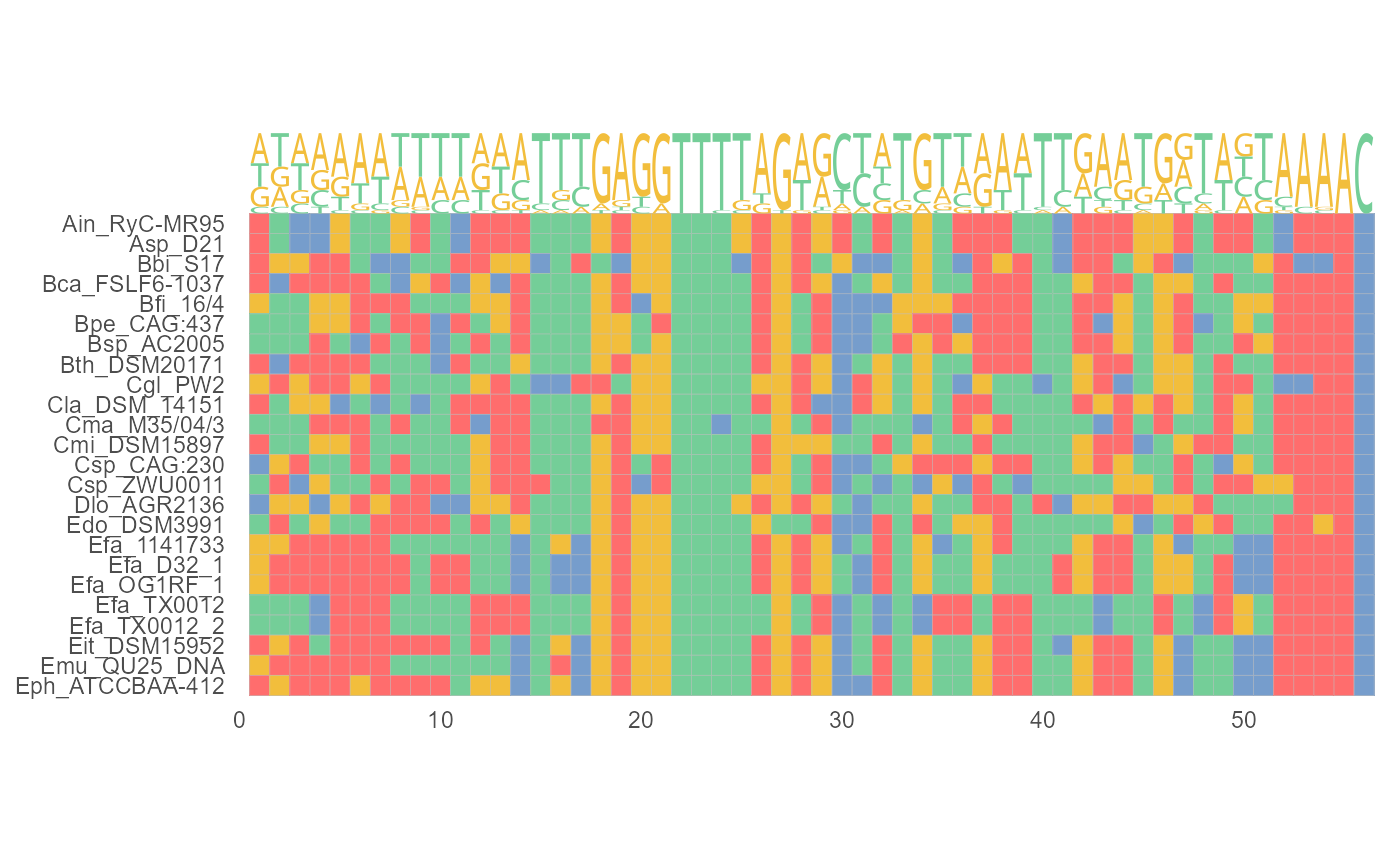
geom_seqlogo
geom_seqlogo.RdMultiple sequence alignment layer for ggplot2. It plot sequence motifs.
geom_seqlogo( font = "DroidSansMono", color = "Chemistry_AA", adaptive = TRUE, top = TRUE, custom_color = NULL, show.legend = FALSE, ... )
Arguments
| font | font families, possible values are 'helvetical', 'mono', and 'DroidSansMono', 'TimesNewRoman'. Defaults is 'DroidSansMono'. |
|---|---|
| color | A Color scheme. One of 'Clustal', 'Chemistry_AA', 'Shapely_AA', 'Zappo_AA', 'Taylor_AA', 'LETTER', 'CN6', 'Chemistry_NT', 'Shapely_NT', 'Zappo_NT', 'Taylor_NT'. Defaults is 'Chemistry_AA'. |
| adaptive | A logical value indicating whether the overall height of seqlogo corresponds to the number of sequences.If is FALSE, seqlogo overall height = 4,fixedly. |
| top | A logical value. If TRUE, seqlogo is aligned to the top of MSA. |
| custom_color | A data frame with two cloumn called "names" and "color".Customize the color scheme. |
| show.legend | logical. Should this layer be included in the legends? |
| ... | additional parameter |
Value
A list
Author
Lang Zhou
Examples
#plot multiple sequence alignment and sequence motifs f <- system.file("extdata/LeaderRepeat_All.fa", package="ggmsa") ggmsa(f,font = NULL,color = "Chemistry_NT") + geom_seqlogo()