View Modes
2021-09-22
View_modes.Rmdggmsa also uses functional views to make MSA features prominent. For instance, for specific properties that predominate regions, there are two methods to highlight them.
Consensus Views
Using consensus_views option to executes Consensus View. We provide four parameters——use_dot, disagreement, ignore_gaps and ref to adjust the Consensus View executes.
-
use_dot: a logical value. Displays characters as dots instead of fading their color in the consensus view. -
disagreement: a logical value. Displays characters that disagreement with consensus (excludes ambiguous disagreements). -
ignore_gaps: a logical value. When selected TRUE, gaps in the column are treated as if that row didn’t exist. -
ref: a character string. Specifying the reference sequence which should be one of the input sequences.
Example 1
When disagreement = TRUE and use_dot = TRUE, characters that agreememt to consensus have displayed as dot.
ggmsa(protein_sequences, 300, 350, char_width = 0.5, seq_name = TRUE, consensus_views = TRUE, disagreement = TRUE, use_dot = TRUE)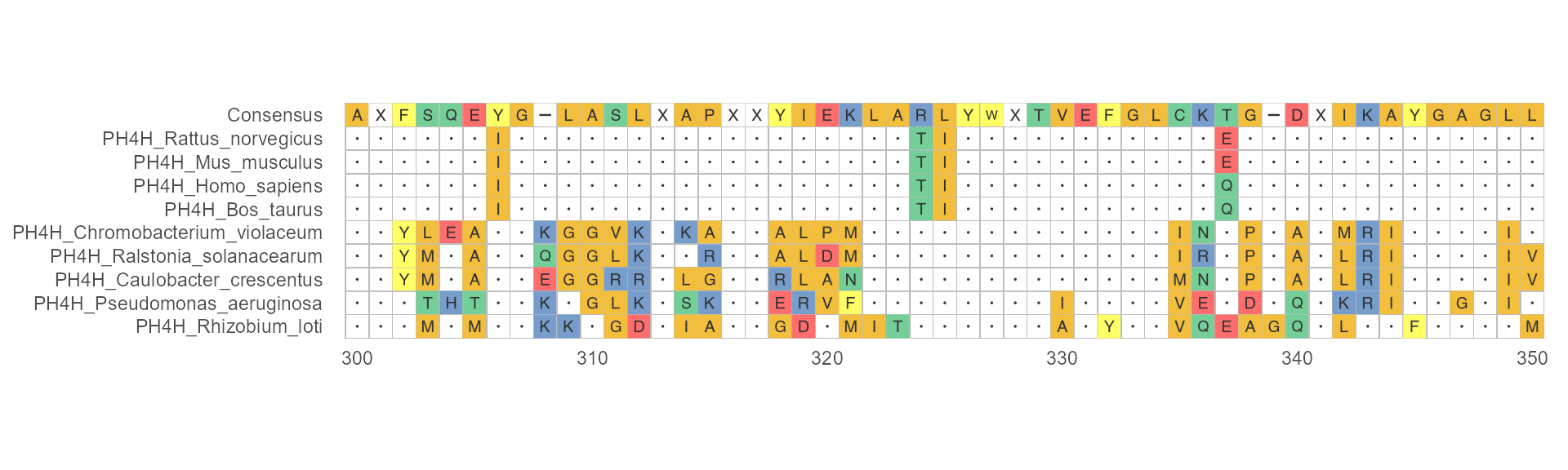
Example 2
When disagreement = FALSE and use_dot = FALSE, characters that disagreement to consensus have faded their colors.
ggmsa(protein_sequences, 300, 350, char_width = 0.5, seq_name = TRUE, consensus_views = TRUE, disagreement = FALSE, use_dot = FALSE)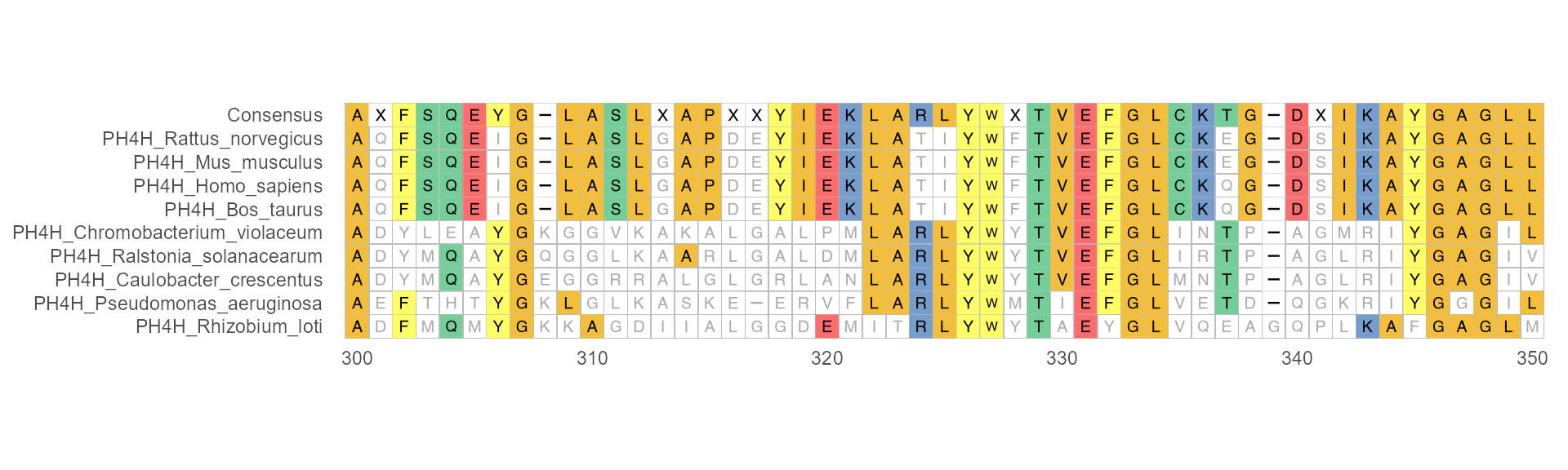
Example 3
Specifying one of the input sequences as the reference sequence. The reference sequence will be placed at the top.
ggmsa(protein_sequences, 300, 350, char_width = 0.5, seq_name = TRUE, consensus_views = TRUE ,use_dot = TRUE, ref = "PH4H_Rhizobium_loti")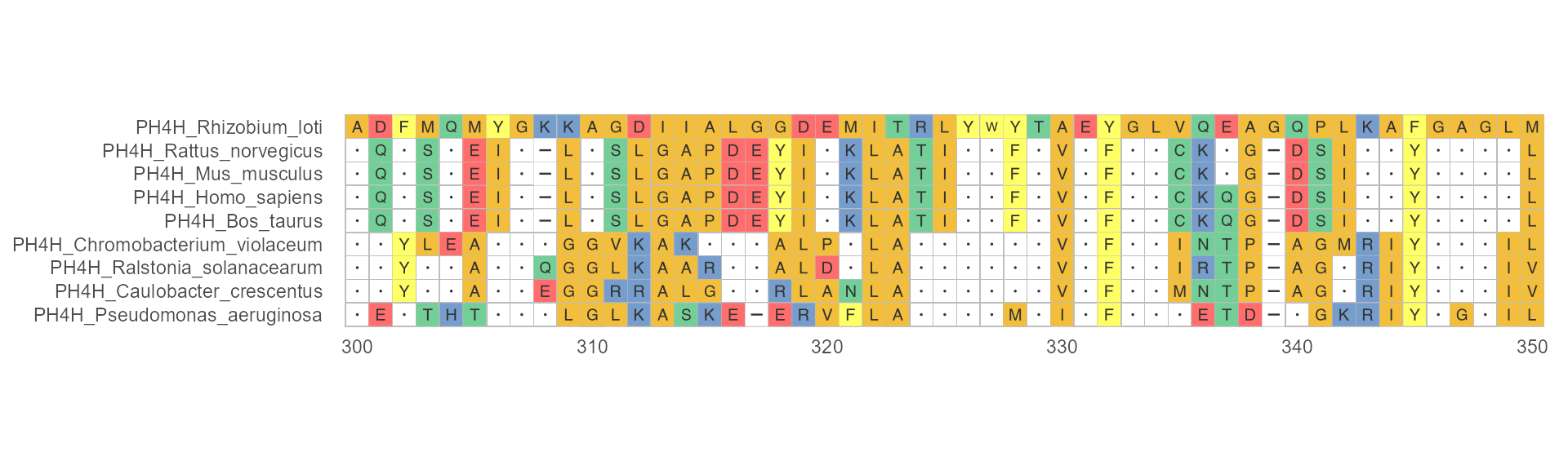
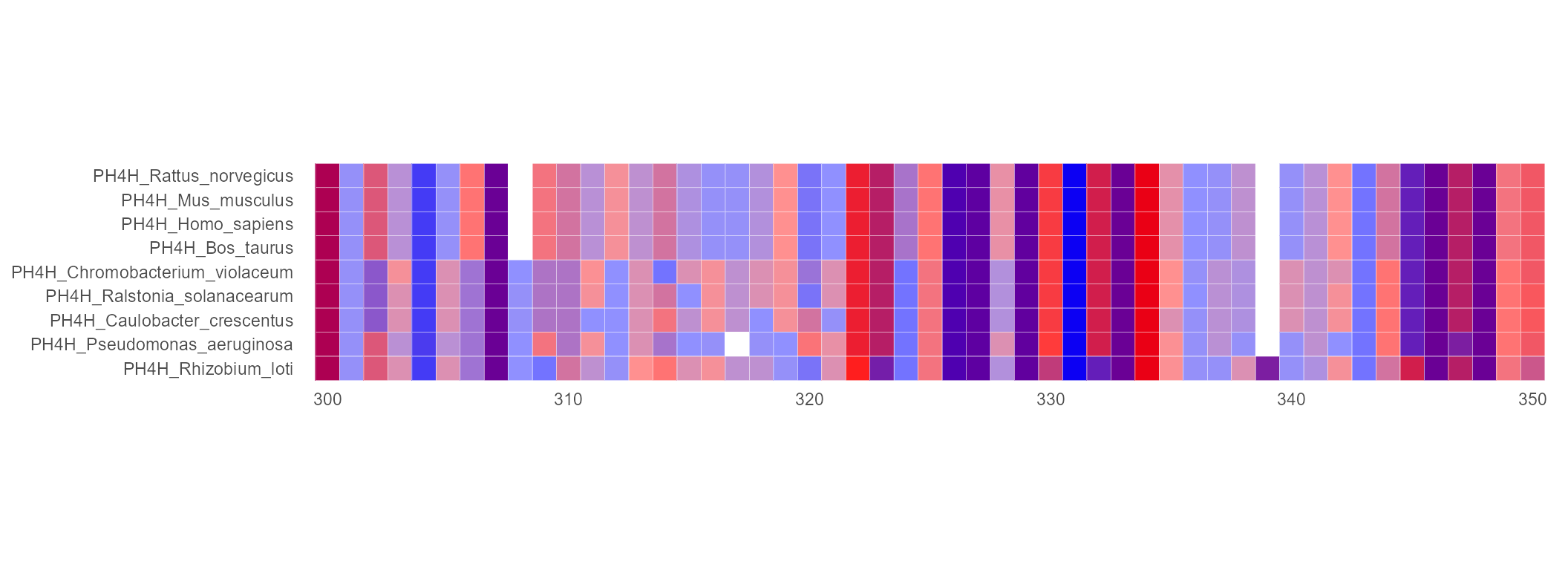
By Conservation
The existing color scheme is modified so that the most conserved position in each column has the most intense colors and the least conserved are the palest. For example, conserved hydrophobic columns are shown with predominately red residues and conserved hydrophilic columns with blue in the Hydrophobicity color scheme.
ggmsa(protein_sequences, 300, 350, color = "Hydrophobicity",
font = NULL, seq_name = TRUE ,border = "white", by_conservation = TRUE)