Chapter 9 MeSH Enrichment Analysis
meshes supports enrichment analysis (over-representation analysis and gene set
enrichment analysis) of gene list or whole expression profile using MeSH
annotation. Data source from gendoo, gene2pubmed and RBBH are all
supported. User can selecte interesting category to test. All 16
categories are supported. The analysis supports >70 species listed in MeSHDb BiocView.
For algorithm details, please refer to the vignettes of DOSE(Yu et al. 2015) package.
library(meshes)
data(geneList, package="DOSE")
de <- names(geneList)[1:100]
x <- enrichMeSH(de, MeSHDb = "MeSH.Hsa.eg.db", database='gendoo', category = 'C')
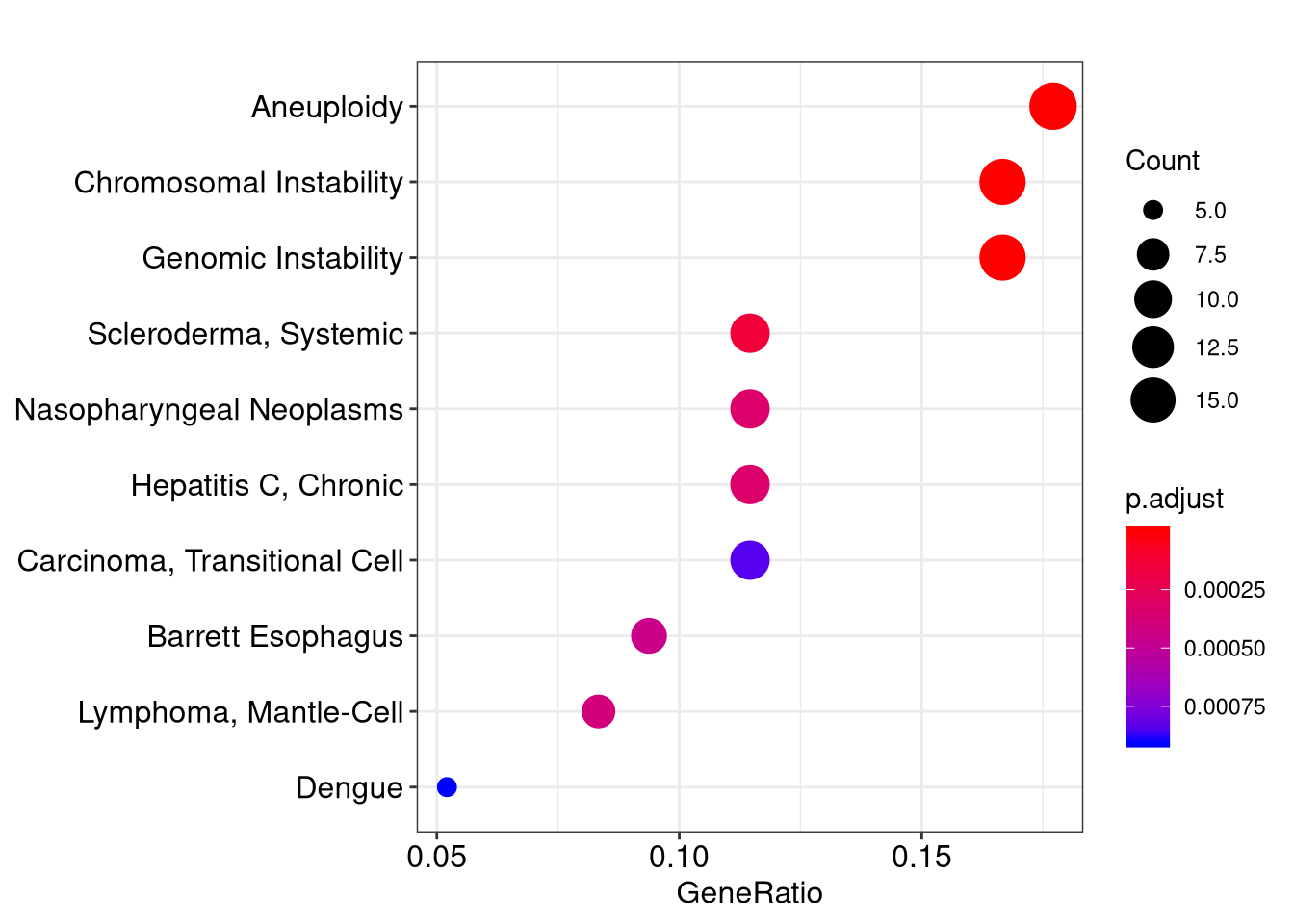
head(x)## ID Description GeneRatio
## D043171 D043171 Chromosomal Instability 16/96
## D000782 D000782 Aneuploidy 17/96
## D042822 D042822 Genomic Instability 16/96
## D012595 D012595 Scleroderma, Systemic 11/96
## D009303 D009303 Nasopharyngeal Neoplasms 11/96
## D019698 D019698 Hepatitis C, Chronic 11/96
## BgRatio pvalue p.adjust qvalue
## D043171 198/16528 2.794765e-14 2.459394e-11 1.815127e-11
## D000782 320/16528 3.866830e-12 1.701405e-09 1.255702e-09
## D042822 312/16528 3.007419e-11 8.821761e-09 6.510798e-09
## D012595 279/16528 6.449334e-07 1.418854e-04 1.047168e-04
## D009303 314/16528 2.049315e-06 3.295389e-04 2.432123e-04
## D019698 317/16528 2.246856e-06 3.295389e-04 2.432123e-04
## geneID
## D043171 4312/991/2305/1062/4605/10403/7153/55355/4751/4085/81620/332/7272/9212/1111/6790
## D000782 4312/55143/991/1062/7153/4751/79019/55839/890/983/4085/332/7272/9212/8208/1111/6790
## D042822 55143/991/1062/4605/7153/1381/9787/4751/10635/890/4085/81620/332/9212/1111/6790
## D012595 4312/6280/1062/4605/7153/3627/4283/6362/7850/3002/4321
## D009303 4312/7153/3627/6241/983/4085/5918/332/3002/4321/6790
## D019698 4312/3627/10563/6373/4283/983/6362/7850/332/3002/3620
## Count
## D043171 16
## D000782 17
## D042822 16
## D012595 11
## D009303 11
## D019698 11In the over-representation analysis, we use data source from gendoo and C (Diseases) category.
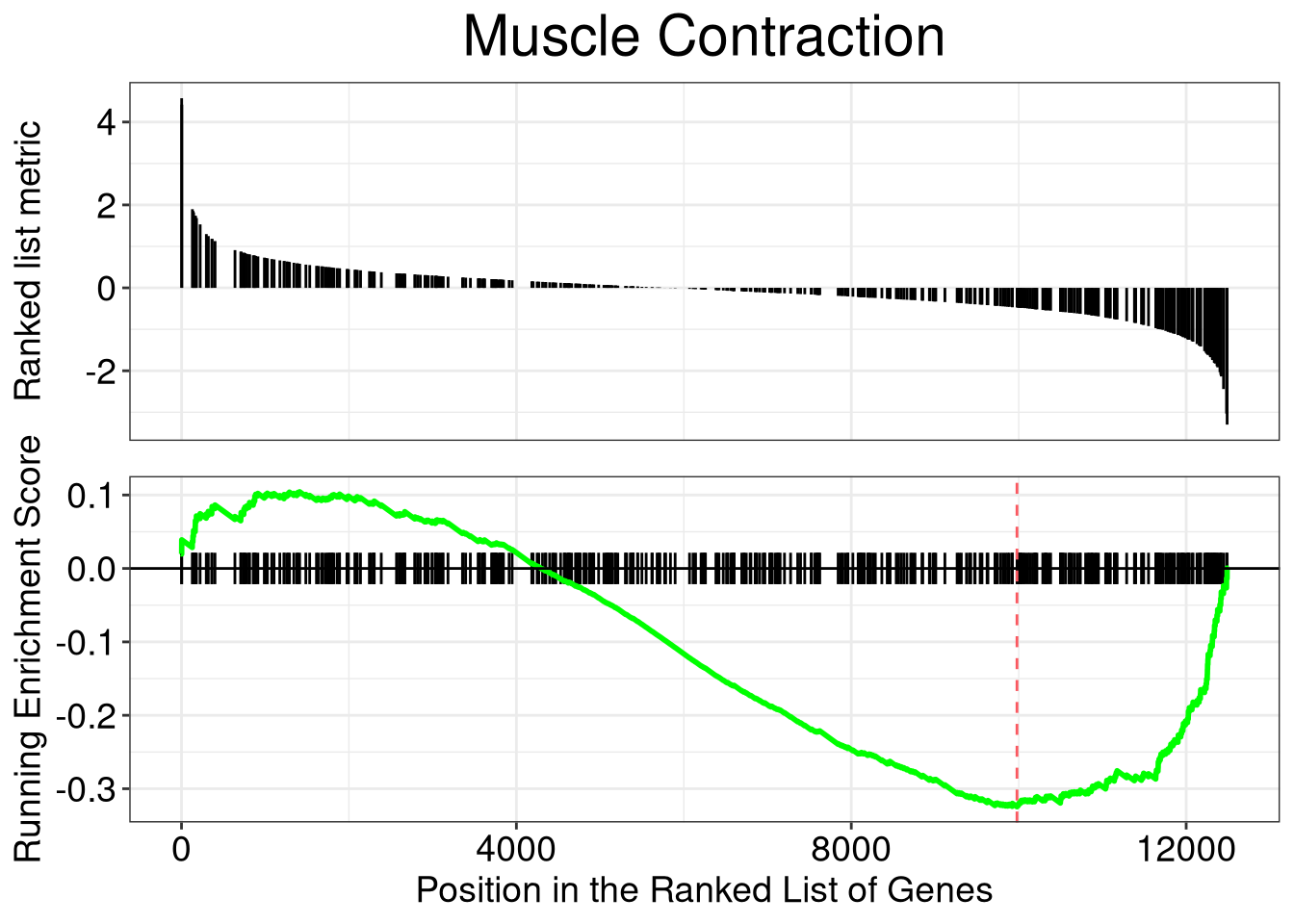
In the following example, we use data source from gene2pubmed and test category G (Phenomena and Processes) using GSEA.
## ID Description setSize enrichmentScore
## D009119 D009119 Muscle Contraction 438 -0.3244845
## D012038 D012038 Regeneration 426 -0.3212385
## D009043 D009043 Motor Activity 462 -0.3223426
## D001846 D001846 Bone Development 322 -0.3722690
## D006339 D006339 Heart Rate 336 -0.3640991
## D049629 D049629 Waist-Hip Ratio 321 -0.3659337
## NES pvalue p.adjust qvalues rank
## D009119 -1.430246 0.001236094 0.03722703 0.0278947 2517
## D012038 -1.413109 0.001237624 0.03722703 0.0278947 2132
## D009043 -1.422254 0.001240695 0.03722703 0.0278947 2176
## D001846 -1.598351 0.001295337 0.03722703 0.0278947 2100
## D006339 -1.566245 0.001300390 0.03722703 0.0278947 2405
## D049629 -1.569546 0.001300390 0.03722703 0.0278947 2176
## leading_edge
## D009119 tags=27%, list=20%, signal=22%
## D012038 tags=27%, list=17%, signal=23%
## D009043 tags=23%, list=17%, signal=20%
## D001846 tags=27%, list=17%, signal=23%
## D006339 tags=29%, list=19%, signal=24%
## D049629 tags=27%, list=17%, signal=23%
## core_enrichment
## D009119 5742/10174/2150/5562/3611/22859/4604/7070/4985/7139/3784/154/1760/3315/9732/72/5595/3092/6416/9759/270/6558/627/953/408/2908/7138/5563/6794/5564/3567/2104/845/3371/6548/831/182/3554/126393/7402/1129/7201/3350/5590/5592/7168/79923/2149/4628/23426/8082/5021/2318/23284/844/79026/4208/3790/2308/1907/253959/54795/4311/2247/10580/1848/2281/10398/5166/50507/1012/6876/10203/83700/11167/2317/3952/3778/1009/5733/10468/3693/6253/9499/7481/5159/3991/857/1289/1909/6678/7041/32/8639/5350/3551/1264/2697/185/55107/7043/3357/2205/253190/5327/25802/1634/3572/8490/3679/3479/5348/9370/9122/4629/652/7021/5241
## D012038 2869/5087/1499/7157/79960/627/2252/4088/825/9149/8038/4017/7010/2752/3248/3082/22921/3791/4005/182/7402/7474/596/947/9976/9315/8840/1490/54209/1280/4804/4314/324/6019/8425/595/10979/6843/4929/79026/2246/5029/4803/7042/4322/8829/7048/10216/79679/5176/55384/7078/5549/7216/727/10516/2247/6591/56944/210/5468/23345/6469/216/8076/26509/90865/11167/7075/7058/4313/3861/91851/2199/113146/6444/9201/1294/4254/4856/6720/3480/5764/6387/6833/5159/11117/857/1289/3908/4016/6678/7033/23030/7704/174/1191/2737/5744/11098/10631/9429/214/7043/2200/1634/4582/7031/3479/7373/2066/3169/2625
## D009043 6532/10550/9759/23405/1499/6453/8945/7157/25970/627/408/2908/22881/27445/11132/2752/9445/6548/2571/23621/3082/1291/2915/1543/7466/3240/3350/947/55304/181/3632/2169/27306/1621/80169/9627/196/8678/8863/23284/81627/4692/5799/11076/2259/3087/1278/283/1277/3953/4747/2247/6414/210/4744/5468/8835/89795/4023/8522/4319/3485/3952/79068/8864/4313/2944/2273/2099/3480/8528/4908/56892/3339/5138/57161/4741/4306/6571/79750/4915/5744/2487/58503/347/6863/2952/5327/367/4982/4128/4059/3572/150/7060/9358/7166/3479/9254/5348/4129/9370/3708/1311/5105/4137/1408/5241
## D001846 1499/8945/7157/57798/79048/627/6500/8038/4057/860/2752/4882/3371/2915/5745/63971/54455/3791/819/57045/596/2034/54808/80781/1280/64388/2261/4054/11059/3483/9900/26234/4734/9452/4208/4322/253461/1278/7048/51280/10903/30008/7869/1277/3953/10516/10411/8835/79776/11167/2317/3485/3952/5274/54681/4488/10486/1009/2202/91851/2099/5764/23327/3339/8817/83716/6678/4915/633/658/54361/5744/165/5654/10631/3487/367/4982/3667/79971/1634/3479/114899/9370/652/8614/4969
## D006339 83478/4985/7139/8929/3784/10681/3375/154/1760/9781/5139/118/2702/6532/6416/2869/270/7157/627/2908/7138/5563/3643/1129/7779/947/1901/2034/4179/4804/64388/1621/4881/8863/5021/844/4212/11030/5797/6403/4803/84059/79789/5176/3953/5243/5468/1012/2868/5793/4023/7056/3952/5577/126/2946/3778/477/5733/4313/2944/9201/3075/9499/2273/2099/1471/857/775/5138/4306/4487/213/5350/5744/23245/2152/2697/2791/185/6863/2952/5327/80206/2200/9607/3572/150/8490/3479/2006/55259/9370/125/652/55351
## D049629 6532/8609/9563/23405/10206/7157/23314/4776/25970/627/2908/490/4057/268/3567/23429/283450/1543/3240/3174/81490/23047/55304/5099/54808/4179/2169/948/8082/4018/54465/4256/3087/5919/253461/26470/10903/1581/56172/3953/5950/5468/1012/8835/4023/594/4214/7350/3952/79068/51232/2202/6444/9369/2099/6833/3991/4016/2690/57161/79750/4915/5125/5167/8639/11188/10631/3551/2487/2697/6935/3487/367/4982/3667/4059/150/9358/1489/3479/6424/9370/4629/652/5346/7021/4239/5241User can use visualization methods implemented in enrichplot (i.e.barplot, dotplot, cnetplot, emapplot and gseaplot) to visualize these enrichment results. With these visualization methods, it’s much easier to interpret enriched results.
References
Yu, Guangchuang, Li-Gen Wang, Guang-Rong Yan, and Qing-Yu He. 2015. “DOSE: An R/Bioconductor Package for Disease Ontology Semantic and Enrichment Analysis.” Bioinformatics 31 (4): 608–9. https://doi.org/10.1093/bioinformatics/btu684.