Chapter 4 🌱 Tumor Microenvironment Analysis
4.1 Availiable TME analysis method in tigeR
10 open source tumor microenvironment deconvolution methods are indlcued in tigeR.
| Algorithm | license | PMID | allow custom signature matrix |
|---|---|---|---|
| TIMER | free (GPL 2.0) | 27549193 | √ |
| CIBERSORT | free for non-commercial use only | 31061481 | √ |
| MCPCounter | free (GPL 3.0) | 27765066 | × |
| xCell | free (GPL 3.0) | 29141660 | × |
| IPS | free (BSD) | 28052254 | × |
| EPIC | free for non-commercial use only (Academic License) | 29130882 | √ |
| ESTIMATE | free (GPL 2.0) | 24113773 | × |
| ABIS | free (GPL 2.0) | 30726743 | √ |
| ConsensusTME | free (GPL 3.0) | 31641033 | × |
| quanTIseq | free (BSD) | 31126321 | × |
4.2 Derive the proportions of different TME cell types
You can use the function deconv_TME() to derive the proportions of different tumor microenvironment cell types from gene expression data with these tools.
## Downloading GitHub repo dviraran/xCell@HEAD## Error in utils::download.file(url, path, method = method, quiet = quiet, :
## cannot open URL 'https://api.github.com/repos/dviraran/xCell/tarball/HEAD'## Skipping install of 'EPIC' from a github remote, the SHA1 (50a4f404) has not changed since last install.
## Use `force = TRUE` to force installation## Downloading GitHub repo cansysbio/ConsensusTME@HEAD## Error in utils::download.file(url, path, method = method, quiet = quiet, :
## cannot open URL 'https://api.github.com/repos/cansysbio/ConsensusTME/tarball/HEAD'## Downloading GitHub repo federicomarini/quantiseqr@HEAD## Error in utils::download.file(url, path, method = method, quiet = quiet, :
## cannot open URL 'https://api.github.com/repos/federicomarini/quantiseqr/tarball/HEAD'## Found 130 genes with uniform expression within a single batch (all zeros); these will not be adjusted for batch.## CIBERSORT
frac2 <- deconv_TME(MEL_GSE78220,method="CIBERSORT")
## MCPCounter
frac3 <- deconv_TME(MEL_GSE78220,method="MCPCounter")
## xCell
frac4 <- deconv_TME(MEL_GSE78220,method="xCell")## [1] "Num. of genes: 10808"
## Setting parallel calculations through a MulticoreParam back-end
## with workers=4 and tasks=100.
## Estimating ssGSEA scores for 489 gene sets.
## [1] "Calculating ranks..."
## [1] "Calculating absolute values from ranks..."
##
|
| | 0%
|
|== | 4%
|
|===== | 7%
|
|======== | 11%
|
|========== | 14%
|
|============ | 18%
|
|=============== | 21%
|
|================== | 25%
|
|==================== | 29%
|
|====================== | 32%
|
|========================= | 36%
|
|============================ | 39%
|
|============================== | 43%
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
|
|======================================== | 57%
|
|========================================== | 61%
|
|============================================= | 64%
|
|================================================ | 68%
|
|================================================== | 71%
|
|==================================================== | 75%
|
|======================================================= | 79%
|
|========================================================== | 82%
|
|============================================================ | 86%
|
|============================================================== | 89%
|
|================================================================= | 93%
|
|==================================================================== | 96%
|
|======================================================================| 100%## IPS
frac5 <- deconv_TME(MEL_GSE78220,method="IPS")
## EPIC
frac6 <- deconv_TME(MEL_GSE78220,method="epic")
## ESTIMATE
frac7 <- deconv_TME(MEL_GSE78220,method="ESTIMATE")## [1] "Merged dataset includes 10130 genes (282 mismatched)."
## [1] "1 gene set: StromalSignature overlap= 137"
## [1] "2 gene set: ImmuneSignature overlap= 141"## ABIS
frac8 <- deconv_TME(MEL_GSE78220,method="ABIS")
## ConsensusTME
frac9 <- deconv_TME(MEL_GSE78220,method="ConsensusTME")## Producing ConsensusTME Estimates Using The Following Parameters:
## Statistical Framework: "ssgsea"
## Gene Sets For Cancer Type: "SKCM"
## Sample Size: 28
## Estimating ssGSEA scores for 19 gene sets.
## [1] "Calculating ranks..."
## [1] "Calculating absolute values from ranks..."
##
|
| | 0%
|
|== | 4%
|
|===== | 7%
|
|======== | 11%
|
|========== | 14%
|
|============ | 18%
|
|=============== | 21%
|
|================== | 25%
|
|==================== | 29%
|
|====================== | 32%
|
|========================= | 36%
|
|============================ | 39%
|
|============================== | 43%
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
|
|======================================== | 57%
|
|========================================== | 61%
|
|============================================= | 64%
|
|================================================ | 68%
|
|================================================== | 71%
|
|==================================================== | 75%
|
|======================================================= | 79%
|
|========================================================== | 82%
|
|============================================================ | 86%
|
|============================================================== | 89%
|
|================================================================= | 93%
|
|==================================================================== | 96%
|
|======================================================================| 100%
##
## [1] "Normalizing..."4.3 Visualization and comparing the cell proportions
## Found 130 genes with uniform expression within a single batch (all zeros); these will not be adjusted for batch.## Found2batches## Adjusting for0covariate(s) or covariate level(s)## Standardizing Data across genes## Fitting L/S model and finding priors## Finding parametric adjustments## Adjusting the Data## CIBERSORT
frac2 <- deconv_TME(MEL_GSE78220,method="CIBERSORT")
cell1 <- c("T cells CD4","Neutrophil", "Macrophage","mDCs","B cells", "T cells CD8")
pie1 <- fraction_pie(cell_name_filter(frac1),feature=factor(cell1, levels = cell1))
cell2 <- c("DCs resting", "T cells CD8", "T cells CD4 naive", "Macrophages M2", "Yd T cells", "Monocytes","Mast cells resting", "Neutrophils", "Tregs","B cells naive")
pie2 <- fraction_pie(cell_name_filter(assay(frac2[1:22,])),
feature=factor(cell2, levels = cell2))
pie1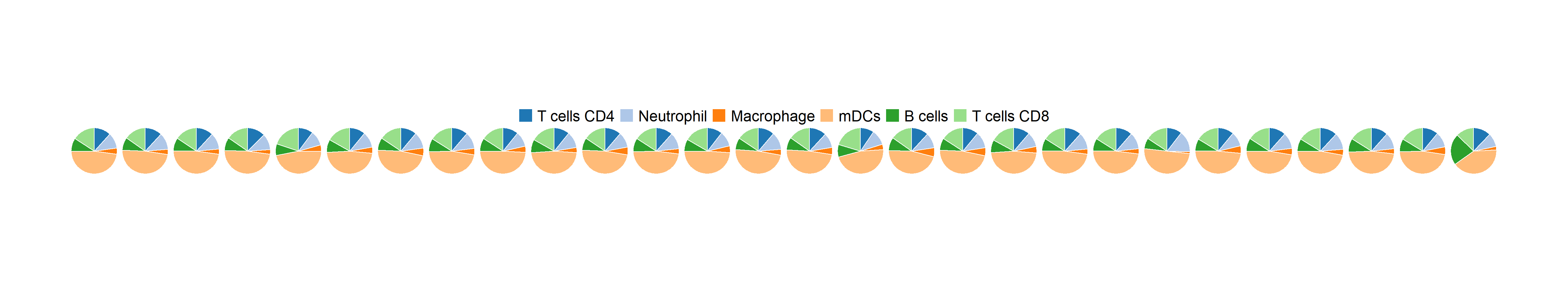
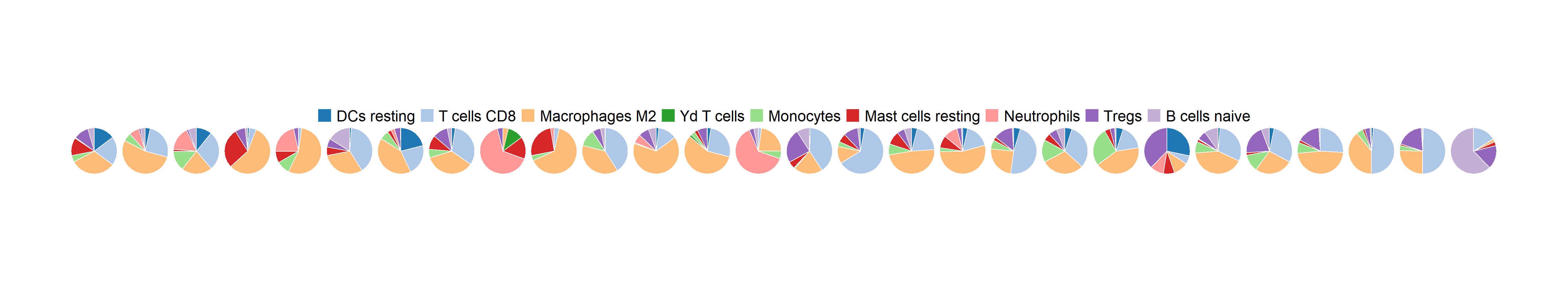
4.4 Searching for key cell types associated with immunotherapy response
## Found 125 genes with uniform expression within a single batch (all zeros); these will not be adjusted for batch.TM_SE <- SummarizedExperiment(assays=SimpleList(TM),
colData=colData(MEL_GSE91061))
browse_biomk(SE=TM_SE)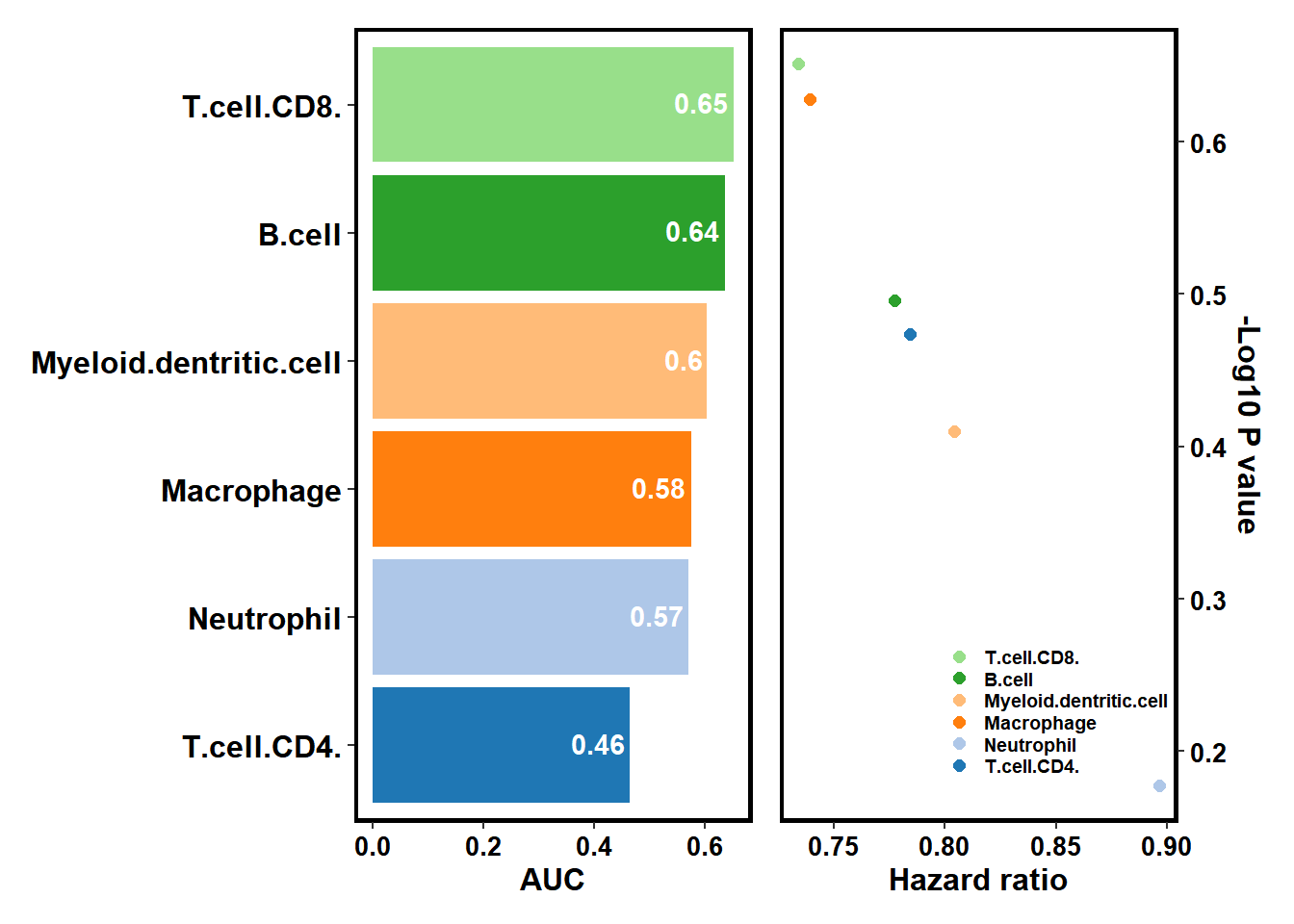
4.5 Generate a custom reference matrix for TME deconvolution from single-cell sequencing data
library(Seurat)
library(tigeR.data)
library(magrittr)
library(GenomicFeatures)
pbmc <- readRDS(system.file("extdata","pbmc.rds",
package = "tigeR",
mustWork = TRUE))
pbmc <- FindVariableFeatures(pbmc,
selection.method = "vst",
nfeatures = 5000, verbose = FALSE,assay = "RNA")
pbmc <- NormalizeData(pbmc,
normalization.method = "LogNormalize",
scale.factor = 10000, verbose = FALSE)
pbmc <- ScaleData(pbmc, features = rownames(pbmc), verbose = FALSE)
pbmc <- RunPCA(pbmc, features = VariableFeatures(object = pbmc), verbose = FALSE)
pbmc <- FindNeighbors(pbmc, dims = 1:10, verbose = FALSE)
pbmc <- FindClusters(pbmc, resolution = 0.5, verbose = FALSE)
new.cluster.ids <- c("Naive_CD4T", "CD14_Mono", "Memory_CD4T", "B_cell", "CD8T", "FCGR3A_Mono", "NK", "DC", "Platelet")
names(new.cluster.ids) <- levels(pbmc$seurat_clusters)
pbmc <- RenameIdents(pbmc, new.cluster.ids)
PCAPlot(pbmc, reduction = "umap", label = TRUE, pt.size = 0.5) + NoLegend()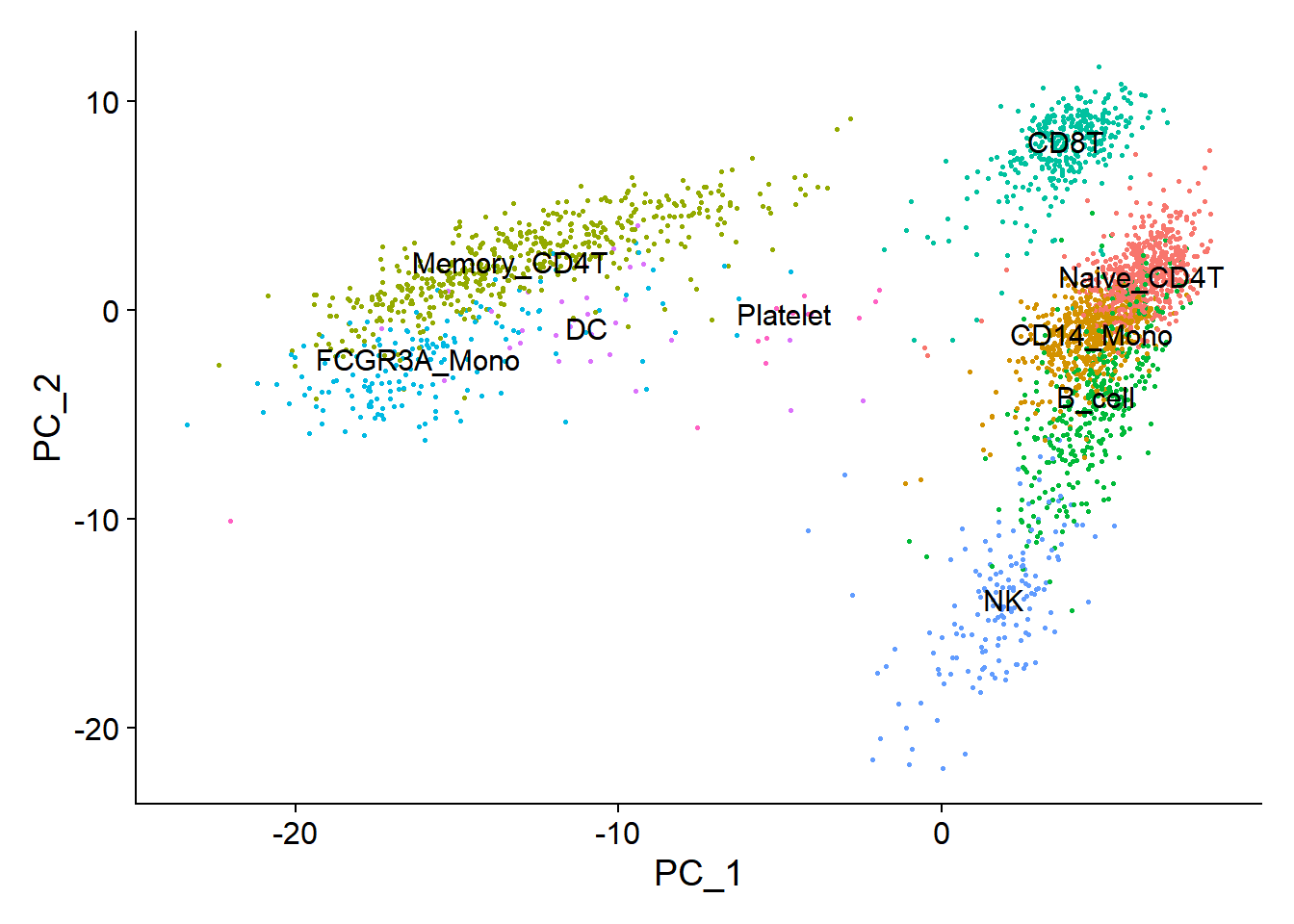
## Naive_CD4T CD14_Mono Memory_CD4T B_cell CD8T
## AAK1 15.0071873 1.062216e+01 1.538455e+00 1.028047e+01 1.677737e+00
## AAMP 35.6795134 3.877492e+01 4.176995e+01 4.147438e+01 9.897730e+01
## ABCF3 4.6785876 7.324110e+00 3.558353e+00 6.056491e+00 1.014087e+01
## ABHD14B 83.7383154 9.681423e+01 2.843133e+01 9.111420e+01 6.139144e+01
## ABHD16A 4.4975028 4.854155e+00 4.247081e+00 3.526542e+00 2.488504e+00
## ABHD5 12.6032567 3.345934e+00 4.867387e+00 1.290599e+00 2.129030e+00
## ACAA1 19.3165000 1.826831e+01 2.368256e+01 2.586982e+01 1.977010e+01
## ACTR2 49.0506904 5.002758e+01 7.170890e+01 5.686290e+01 7.634531e+01
## ACTR3 45.6092143 7.302694e+01 3.317173e+01 6.906637e+01 6.274021e+01
## ADAR 14.6424796 1.761073e+01 1.639854e+01 2.286557e+01 2.216309e+01
## ADH5 37.8990360 6.269716e+01 1.850506e+01 5.261235e+01 5.486930e+01
## ADI1 37.1979283 5.656108e+01 3.490842e+01 2.263481e+01 3.152288e+01
## AFF3 0.9830185 1.173923e-01 3.132282e-01 8.223931e-01 1.426946e+01
## AGPAT1 42.8365647 7.920439e+00 9.305385e+00 1.624317e+01 4.478442e+00
## AGTRAP 24.2272498 3.293684e+01 2.193184e+02 3.660363e+01 2.313783e+01
## AIF1 457.8812896 1.691177e+02 4.554946e+03 1.426525e+02 1.012504e+02
## AKR1A1 43.2777197 4.567830e+01 8.202105e+01 5.394508e+01 5.969339e+01
## AMPD2 4.3767802 4.378425e+00 9.288163e+00 2.800810e+00 3.586318e+00
## ANXA4 2.1107036 8.078455e+00 7.442711e+00 1.285823e+01 2.173704e+01
## ANXA5 72.7211799 1.842670e+02 3.043088e+02 1.152857e+02 8.952911e+01
## ANXA6 63.8066878 1.089558e+02 3.007343e+01 1.446243e+02 1.357757e+02
## ARHGAP15 86.4328209 1.247354e+02 1.600380e+01 8.675396e+01 6.429643e+01
## ARHGAP24 1.0473690 0.000000e+00 2.320084e+01 0.000000e+00 3.686333e+01
## ARHGEF3 9.9566693 1.791160e+01 5.337658e+00 1.515462e+01 8.544247e+00
## ARL4C 83.5857757 9.435623e+01 2.503875e+01 1.379403e+02 2.161817e+01
## ARL6IP5 331.7905146 4.677853e+02 1.849818e+02 5.491067e+02 2.291578e+02
## ARPC2 237.8405218 3.699571e+02 4.536223e+02 4.135716e+02 3.325102e+02
## ARPC4 182.2148346 1.520892e+02 1.603541e+02 2.272185e+02 1.236026e+02
## ARPC5 75.6939236 1.052623e+02 9.813458e+01 8.960859e+01 7.253781e+01
## ARSB 0.0000000 1.536196e-01 1.375768e+00 8.405583e-01 1.628779e+00
## ASAP2 0.0000000 0.000000e+00 1.423681e+00 0.000000e+00 0.000000e+00
## ATF3 5.8411293 7.539935e+00 4.713817e+01 3.838902e+00 7.432398e+00
## ATG3 32.1306930 3.010076e+01 6.701022e+01 4.904289e+01 3.985619e+01
## ATOX1 13.6195025 2.206544e+01 6.003925e+01 1.649238e+01 1.340707e+01
## ATP1A1 44.1599471 4.566071e+01 2.492675e+01 3.877367e+01 1.974459e+01
## ATP1B3 30.6907235 7.540906e+01 7.098092e+01 1.227455e+02 5.913295e+01
## ATP6V0B 69.4935109 1.025100e+02 4.158346e+02 1.049206e+02 1.403196e+02
## BAG6 14.4398770 1.425262e+01 1.090838e+01 1.557428e+01 2.226589e+01
## BANK1 6.5846354 6.253767e+00 6.630983e+00 9.982593e+00 1.863043e+02
## BASP1 1.4752657 1.511741e+00 1.811121e+01 1.087188e+00 3.803765e+01
## BATF3 2.4682847 1.204271e+00 8.647560e+00 0.000000e+00 2.996330e+00
## BCL11A 0.0753446 3.734603e-01 1.048506e+00 0.000000e+00 4.920621e+00
## BHLHE40 7.1077185 1.921726e+01 2.401753e+01 2.927377e+01 9.738793e+00
## BIN1 70.1289706 8.558018e+01 6.727732e+00 7.922653e+01 8.155756e+01
## BRK1 627.6039431 7.588635e+02 1.127809e+03 9.970553e+02 9.939457e+02
## BST1 5.3941297 4.751016e+00 1.294412e+02 1.130816e+00 1.498910e+00
## BTG2 313.6343158 3.233706e+02 1.037568e+02 2.170296e+02 2.136382e+02
## BTN3A2 48.0155733 4.899579e+01 2.242047e+01 7.168576e+01 8.133021e+01
## BZW1 27.6980799 2.712524e+01 1.131470e+01 4.283888e+01 2.649571e+01
## C1orf122 31.4696567 3.939486e+01 3.375084e+01 4.815441e+01 4.107986e+01
## C1orf162 172.8588364 1.632392e+02 5.484153e+02 7.509812e+01 1.623852e+02
## C1orf198 0.4035237 4.114592e-01 2.045716e-01 8.449740e-01 0.000000e+00
## C1orf21 0.1632966 0.000000e+00 1.005080e+00 1.178623e+01 7.108623e-01
## C1orf43 84.6242180 1.213803e+02 1.275234e+02 1.017564e+02 8.260642e+01
## C1QA 1.6407988 0.000000e+00 1.579291e+01 1.314992e+00 0.000000e+00
## C2orf88 0.0000000 1.791027e-01 4.016476e-01 0.000000e+00 0.000000e+00
## CACNA2D3 0.0000000 3.104931e-01 1.321690e+01 0.000000e+00 0.000000e+00
## CALM2 104.0935295 1.489917e+02 1.608754e+02 1.546537e+02 1.440122e+02
## CAMK1 5.1385221 5.965689e+00 5.952546e+01 5.849634e+00 3.184579e+00
## CAMK4 19.6508513 1.224008e+01 2.527984e+00 7.627708e+00 5.049599e+00
## CAP1 218.3051921 1.706413e+02 1.460284e+02 2.511547e+02 1.781641e+02
## CAPG 39.3830547 1.088098e+02 3.375149e+02 4.364845e+01 1.952299e+02
## CAPN2 28.5555141 4.817184e+01 2.114386e+01 5.952023e+01 1.787665e+01
## CAPZB 53.0372851 7.104240e+01 4.749356e+01 5.981580e+01 4.320636e+01
## CAST 7.4060993 1.510926e+01 2.023173e+01 1.915414e+01 1.472178e+01
## CCDC12 67.4409356 6.249868e+01 4.393620e+01 7.949728e+01 9.987554e+01
## CCDC167 163.8547268 4.033204e+02 7.167999e+01 4.912101e+02 8.240025e+01
## CCDC28B 12.6639149 8.763086e+00 7.687934e+00 1.638403e+01 1.236732e+01
## CCDC50 1.1540901 2.337504e+00 9.290988e+00 8.259772e-01 2.607771e+01
## CCDC88A 0.4232139 2.886424e-01 1.990636e+00 6.307689e-01 9.144578e-01
## CCND3 153.2273529 1.643647e+02 5.129080e+01 1.855735e+02 1.002770e+02
## CCNL1 58.4784983 5.705004e+01 2.515486e+01 4.880900e+01 4.375895e+01
## CCR2 2.0958233 7.511757e+00 7.058539e+00 8.509155e+00 1.197312e+00
## CCT5 32.5965745 4.590272e+01 2.746817e+01 3.874687e+01 4.438749e+01
## CD14 16.7752077 1.312434e+01 9.252201e+02 1.181822e+01 1.965496e+00
## CD160 1.6308510 6.760822e-01 1.506686e+00 5.226628e+01 3.880605e+00
## CD180 0.0000000 8.368661e-01 2.414016e+00 1.408074e+00 4.637501e+01
## CD1C 3.0564186 1.378129e+00 8.241721e+00 6.969663e-01 7.227487e+01
## CD1D 0.7438792 1.091349e+00 4.627847e+01 1.301152e+00 2.211540e+01
## CD2 533.5469951 8.415261e+02 2.054791e+01 7.981971e+02 3.531994e+01
## CD244 5.1710572 8.623657e+00 1.416306e+01 1.946521e+01 2.600401e+00
## CD247 55.0741984 5.753008e+01 3.148277e+00 9.013793e+01 5.364881e+00
## CD302 3.1096864 1.862820e+00 5.012153e+01 4.339830e-01 3.845853e+00
## CD38 3.2636025 2.888329e+00 2.148190e+00 1.710494e+00 9.739753e+00
## CD47 47.9853447 6.030920e+01 2.089141e+01 5.470582e+01 4.844539e+01
## CD48 617.0491082 6.052326e+02 4.725488e+02 5.182746e+02 4.622027e+02
## CD52 4348.1490050 6.112753e+03 3.406237e+03 5.019591e+03 7.180548e+03
## CD53 445.2215834 5.618146e+02 1.707532e+02 6.998478e+02 8.086483e+02
## CD55 27.6213406 2.019485e+01 2.920801e+01 1.261659e+01 3.820053e+01
## CD74 354.0887557 4.541670e+02 1.964214e+03 7.477286e+02 9.459805e+03
## CD83 11.0510014 1.160341e+01 7.221531e+01 1.168832e+01 7.360508e+01
## CD86 2.3723376 2.146693e+00 4.336098e+01 2.373432e+00 3.128821e+00
## CD8A 53.7883142 2.405998e+01 4.458786e+00 2.904781e+02 9.436632e+00
## CD8B 110.8072014 3.736436e+01 2.331970e+00 2.187097e+02 4.973373e+00
## CD96 34.6218063 4.375871e+01 6.014582e+00 6.066301e+01 1.425724e+01
## CDA 21.9786988 2.799314e+01 1.169320e+03 1.827526e+01 1.060985e+01
## CDC23 4.1373137 4.603074e+00 7.120448e-01 6.238603e+00 3.551099e+00
## CDC42SE1 64.8043470 8.489461e+01 5.297888e+01 8.457350e+01 8.188391e+01
## CDKN1A 9.3817299 1.241406e+01 7.240711e+01 1.559487e+01 1.517060e+01
## CDYL 3.0255824 1.293364e+00 5.562033e+00 4.597776e+00 3.357968e+00
## CEP170 1.5964521 1.183259e+00 1.648125e+00 1.306140e+00 3.497206e+00
## CHCHD5 37.0892026 4.098465e+01 6.968887e+01 5.002234e+01 4.074959e+01
## CHST2 2.4792822 7.838885e-01 2.646714e+00 3.155538e+00 1.180769e+01
## CKS1B 28.8656057 2.935853e+01 1.586680e+01 3.578470e+01 1.359083e+02
## CLIC1 263.7482337 7.335420e+02 9.989081e+02 1.083304e+03 9.732146e+02
## CLIC4 0.3863834 2.613083e-01 4.711875e+00 9.665929e-01 1.792226e+01
## CLTB 70.8007562 1.334020e+02 1.679455e+02 1.294288e+02 8.803414e+01
## CMC1 6.2554860 1.002313e+01 6.287626e+00 3.106874e+01 6.783842e+00
## CMPK2 17.6658494 1.232315e+01 2.542016e+01 7.761012e+00 7.911074e+00
## CMTM6 38.7709152 6.779434e+01 4.747593e+01 4.834341e+01 5.008895e+01
## CMTM7 69.1605894 1.142343e+02 6.599365e+01 1.032128e+02 6.153788e+01
## CMTM8 65.1982155 1.266009e+02 6.483790e+00 2.566707e+01 3.804750e+00
## CNIH4 13.1443623 2.038332e+01 1.546604e+01 2.065821e+01 1.647747e+01
## CNPY3 144.5734617 1.612297e+02 6.646620e+02 2.438878e+02 2.643553e+02
## COA5 27.2148090 2.395186e+01 2.917835e+01 4.062204e+01 3.374499e+01
## COL9A2 0.5693389 0.000000e+00 4.540730e+00 0.000000e+00 2.762883e+00
## COQ2 4.6536182 3.985628e+00 2.709302e+01 8.409969e+00 1.691230e+01
## COX17 106.5873352 1.159043e+02 1.767724e+02 1.127146e+02 1.299765e+02
## COX5B 481.6230607 6.111911e+02 8.458328e+02 4.950277e+02 4.695909e+02
## COX6A1P2 575.3127950 7.608745e+02 6.185901e+02 9.148813e+02 5.523007e+02
## CREG1 11.4798518 7.989735e+00 7.058764e+01 9.227757e+00 1.325875e+01
## CRTAP 5.6212412 1.110599e+01 2.962300e+01 3.625821e+01 1.169044e+01
## CSF1R 3.4367586 1.468661e+00 3.330105e+01 2.003859e+00 3.478663e+00
## CSF3R 3.4250796 3.359994e+00 6.841202e+01 1.177854e+00 1.846913e+00
## CSNK2B 106.9274628 1.385970e+02 1.441357e+02 1.427965e+02 1.284493e+02
## CSRP1 8.1540916 8.156615e+00 5.081100e+00 6.813288e+00 5.014509e+00
## CSTA 1.4813048 1.698477e+00 1.553319e+02 0.000000e+00 1.147734e+00
## CTBS 7.3907262 1.080959e+01 1.180206e+01 6.386847e+00 1.641009e+01
## CTDSP1 30.6233356 3.124174e+01 2.453850e+01 2.895605e+01 2.434572e+01
## CTDSPL 0.7834416 0.000000e+00 5.579283e+00 0.000000e+00 0.000000e+00
## CTNNBIP1 11.3626302 1.324896e+01 3.944295e+01 7.729210e+01 8.534287e+00
## CTSS 80.9289560 7.798898e+01 7.318379e+02 7.082082e+01 1.743382e+02
## CX3CR1 7.8331518 4.042259e+00 3.608091e+01 4.197884e+01 8.260161e+00
## CXCR4 807.6470643 8.325294e+02 1.054065e+02 7.783854e+02 1.395768e+03
## CXXC5 3.8723932 6.138494e+00 1.816573e+01 2.868779e+01 9.605186e+01
## CYB5R1 15.3930435 1.244788e+01 9.970400e+00 1.791026e+01 1.174741e+01
## CYSTM1 7.0936046 2.461970e+01 7.228520e+01 2.521462e+01 2.345422e+01
## CYTIP 226.6880938 2.014300e+02 6.831846e+01 1.609604e+02 1.897345e+02
## DAB2 0.0000000 0.000000e+00 1.031365e+00 9.947693e-01 0.000000e+00
## DAPP1 12.9103337 1.363663e+01 1.679805e+01 5.491195e+00 4.212070e+01
## DBI 134.0958418 1.838037e+02 2.036871e+02 2.555575e+02 3.438900e+02
## DDAH2 8.7911690 1.636586e+01 3.400447e+01 3.744132e+01 6.436701e+01
## DDOST 78.5090821 8.928476e+01 7.762991e+01 1.214768e+02 8.281875e+01
## DEF6 43.9727640 4.590656e+01 1.868322e+01 8.495098e+01 2.203349e+01
## DEGS1 70.4481301 1.346216e+02 2.296285e+01 6.931192e+01 9.295127e+01
## DHX29 14.0717165 1.806648e+01 7.391118e+00 1.472972e+01 1.623564e+01
## DNAJB11 9.6868542 1.169378e+01 1.320106e+01 1.662289e+01 7.697996e+00
## DNASE1L3 0.6454544 0.000000e+00 4.040436e-01 0.000000e+00 0.000000e+00
## DOCK2 5.2379406 5.097478e+00 8.615518e+00 1.109412e+01 5.419015e+00
## DOK3 1.9491009 1.579390e+00 5.474202e+01 3.164114e+00 7.944554e+00
## DRAM2 76.3143455 4.853397e+01 6.876569e+01 4.539182e+01 1.333795e+02
## DUSP2 242.4072113 3.469748e+02 8.069210e+01 6.001536e+02 1.153223e+02
## DUSP23 76.9053316 1.115572e+02 2.037815e+02 1.500340e+02 7.208712e+01
## DYNC1LI1 7.2006098 9.440671e+00 5.981240e+00 9.900774e+00 1.046536e+01
## EAF2 16.1362929 1.653088e+01 4.680146e+01 1.649680e+01 4.522369e+02
## EFHD2 23.8137940 4.049720e+01 2.056675e+02 1.029489e+02 4.459201e+01
## EHD3 0.3827974 0.000000e+00 2.976728e-01 1.840116e+00 2.885117e+00
## EIF4A2 335.4777086 3.181476e+02 7.357103e+01 2.777652e+02 3.462473e+02
## EIF4E2 10.6945272 1.382263e+01 2.721647e+01 1.173791e+01 8.107328e+00
## ENO1 210.8690891 2.312778e+02 2.193233e+02 2.520138e+02 1.456842e+02
## ENSA 77.5654970 1.086498e+02 7.620019e+01 9.625598e+01 1.321142e+02
## EREG 0.2082171 3.024439e-01 1.558334e+01 2.441679e+00 6.392927e-01
## F11R 1.0370636 9.750696e-01 3.420540e+00 3.472374e+00 1.518594e+00
## F13A1 2.0328466 1.325932e+00 4.710403e+00 2.582862e+00 2.623422e+00
## FAM76A 5.4264774 3.743353e+00 5.988263e-01 5.158413e+00 6.785792e+00
## FASLG 0.0000000 3.862211e+00 0.000000e+00 1.080439e+01 0.000000e+00
## FAXDC2 0.0000000 5.066628e-01 3.076667e-01 0.000000e+00 0.000000e+00
## FBLN2 0.8935764 2.933823e-01 0.000000e+00 0.000000e+00 8.542414e-01
## FBXO6 4.5875867 1.500010e+01 3.622263e+01 4.841549e+01 3.344309e+01
## FCER1A 31.8333634 1.915435e+00 3.614794e+01 1.125161e+01 2.279700e+00
## FCER1G 130.1428696 1.285080e+02 4.398946e+03 2.903082e+02 1.022386e+02
## FCGR1A 1.0657335 7.239419e-01 1.276479e+02 0.000000e+00 1.968010e+00
## FCGR2A 1.7221793 3.185507e+00 7.591849e+01 0.000000e+00 2.638103e+00
## FCGR2B 1.0891300 9.401111e-01 2.968871e+00 1.602669e+00 4.288296e+01
## FCGR3A 11.0919002 1.361248e+01 3.438358e+01 9.781552e+01 9.861582e+00
## FCRL1 4.0885879 5.980411e+00 3.857720e+00 5.314945e+00 1.067049e+02
## FCRL2 0.8297046 1.163932e+00 3.865316e-01 0.000000e+00 1.352608e+02
## FCRL6 2.0565792 4.823968e+00 5.540139e-01 9.408135e+01 2.112272e+00
## FCRLA 1.2399680 4.202892e+00 4.812744e-01 0.000000e+00 2.114339e+02
## FDPS 42.6626460 4.933385e+01 2.338326e+01 5.691570e+01 6.408421e+01
## FGD2 0.2739118 1.250643e+00 6.914510e+00 1.040651e+00 1.202751e+01
## FGFBP2 17.9865103 2.936288e+01 1.736627e+01 8.544103e+02 4.668457e+01
## FGR 11.9287049 1.408436e+01 2.103665e+02 5.009242e+01 6.529523e+01
## FHIT 52.7414409 1.475157e+01 3.169304e+00 2.306149e+00 6.649637e+00
## FLOT1 43.0070563 4.191950e+01 4.983667e+01 8.597170e+01 2.563091e+01
## FRMD4B 0.9762951 1.303911e+00 1.236671e+00 1.027240e+00 8.278468e-01
## FSTL1 0.3235906 0.000000e+00 3.132564e-01 7.250905e-01 0.000000e+00
## G0S2 28.4467651 3.234248e+01 6.268711e+02 1.227236e+01 1.776858e+01
## GAPT 3.2724713 1.307976e+00 3.811177e+01 2.890349e+00 1.275790e+02
## GATA2 0.5054967 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## GBP2 32.7683449 4.607155e+01 2.762197e+01 5.975107e+01 1.363373e+01
## GCA 18.5057272 1.546759e+01 8.238593e+01 7.586804e+00 4.452136e+01
## GCLM 4.7405443 1.712466e+01 2.116260e+00 2.054533e+01 7.244182e+00
## GLRX 73.8434966 7.214188e+01 1.821672e+02 9.204867e+01 3.515542e+01
## GLUL 4.9541060 8.524236e+00 2.489385e+01 7.502161e+00 7.715310e+00
## GMPR 2.1237256 8.545699e-01 5.067274e+01 0.000000e+00 2.756568e+00
## GNAI2 21.4842996 3.213834e+01 8.248314e+01 5.162596e+01 2.537632e+01
## GNAI3 13.6649965 2.147518e+01 1.203917e+01 2.720899e+01 2.190206e+01
## GNB1 9.4783381 1.168688e+01 2.101539e+01 1.996635e+01 1.016216e+01
## GNB4 0.2877317 2.595648e-01 1.435913e+00 1.137941e+00 4.522854e+00
## GNG5 65.7800564 6.541537e+01 1.142484e+02 7.035712e+01 6.596096e+01
## GNL2 8.2048054 8.585969e+00 8.836509e+00 1.173570e+01 1.163972e+01
## GNLY 39.4654069 4.677173e+01 5.261782e+01 1.259555e+03 4.685106e+01
## GP9 0.0000000 3.588991e+00 1.922764e+00 0.000000e+00 0.000000e+00
## GPBAR1 4.8851172 1.044517e+00 9.577466e+01 2.465498e+00 1.083817e+00
## GPR137B 5.6370394 1.492192e+01 9.583960e+00 1.761048e+01 5.343513e+00
## GPX1 287.5295857 3.258493e+02 2.706934e+03 2.387168e+02 4.832450e+02
## GSTM4 0.4098025 2.219068e+00 2.246517e+00 0.000000e+00 2.591409e+00
## GTPBP2 0.5582234 1.820496e+00 4.350191e+00 0.000000e+00 3.460442e+00
## GYPC 218.1737924 1.418149e+02 3.199992e+01 1.650707e+02 8.754277e+01
## GZMA 90.5162760 2.337342e+02 4.611400e+01 3.308268e+03 2.493123e+01
## GZMK 50.0442399 1.273654e+02 1.141702e+01 1.972866e+03 3.885551e+01
## HAVCR2 1.0714496 1.496372e+00 1.326881e+01 3.560906e+00 1.510299e+00
## HCLS1 80.3721414 1.302076e+02 1.072844e+02 1.194457e+02 1.172670e+02
## HDLBP 3.4405663 4.083089e+00 1.060257e+01 3.891415e+00 5.056754e+00
## HENMT1 19.7280131 3.768822e+01 4.963010e+00 6.067308e+01 1.156893e+01
## HES1 1.0132539 3.046676e+00 1.444230e+01 0.000000e+00 1.416647e+01
## HES4 7.0014940 4.126258e+00 5.684321e+01 0.000000e+00 1.761802e+01
## HGD 0.0000000 1.436334e+00 0.000000e+00 0.000000e+00 0.000000e+00
## HIGD2A 1269.5870966 1.215268e+03 1.758346e+03 1.010131e+03 1.056296e+03
## HINT1 292.5233596 3.290629e+02 8.335156e+01 2.049347e+02 2.998257e+02
## HK3 2.8414730 5.656930e-01 2.454677e+01 3.019053e+00 1.513886e+00
## HLA-DMA 25.5942047 2.129376e+01 4.326329e+02 7.315127e+01 9.845946e+02
## HLA-DMB 11.0356225 9.033112e+00 9.524651e+01 1.973023e+01 3.124223e+02
## HLA-DOA 2.1695848 0.000000e+00 1.687183e+00 2.231776e+00 3.729246e+01
## HLA-DOB 3.4768088 6.411118e+00 4.253557e+00 3.746079e+00 2.871852e+02
## HLA-DPA1 43.0455825 6.627807e+01 6.165388e+02 1.455711e+02 1.485206e+03
## HLA-DPB1 44.5152119 7.130154e+01 5.667724e+02 1.943443e+02 1.752443e+03
## HLA-DQA1 3.2815979 6.337557e+00 6.478983e+01 2.419518e+01 6.558546e+02
## HLA-DQA2 12.6884971 4.595638e+00 1.021693e+02 4.308401e+01 9.386015e+02
## HLA-DQB1 6.9694198 3.311609e+01 7.897264e+01 3.123552e+01 8.997673e+02
## HLA-DQB2 1.8922352 4.034045e+00 3.097430e+00 0.000000e+00 9.900818e+01
## HLA-DRA 325.7209041 4.140921e+02 7.041176e+03 5.351107e+02 2.205666e+04
## HLA-DRB1 45.2396076 9.504795e+01 1.210104e+03 2.668902e+02 2.776985e+03
## HLA-DRB5 40.8960623 7.659385e+01 1.365465e+03 3.233041e+02 3.026303e+03
## HLX 0.0000000 0.000000e+00 3.134608e+00 0.000000e+00 0.000000e+00
## HMGB2 168.5374376 2.446775e+02 2.681517e+02 2.528875e+02 1.578464e+02
## HNMT 3.4239922 8.284963e-01 3.723046e+01 1.836903e+00 1.917916e+00
## HNRNPA3 67.9568967 7.854435e+01 5.366256e+01 1.077961e+02 6.566639e+01
## HNRNPD 17.0737790 2.298853e+01 1.651134e+01 1.962604e+01 1.874406e+01
## HOPX 6.5410800 3.838928e+01 2.109332e+00 1.418849e+02 2.279413e+01
## HRH2 3.7227189 3.945598e+00 1.250525e+01 7.092659e-01 0.000000e+00
## HSD17B11 178.7806506 1.312625e+02 2.521601e+02 1.316195e+02 1.877806e+02
## HSPE1 201.0676013 2.298781e+02 5.795109e+01 2.188679e+02 1.553004e+02
## ID2 244.9635575 3.473455e+02 4.499497e+02 7.240607e+02 5.257812e+01
## IFI44 58.4915177 7.654732e+01 8.746424e+01 1.005320e+02 3.864123e+01
## IFI6 613.3443820 9.155045e+02 2.256865e+03 6.805939e+02 3.431079e+02
## IFT57 24.2665536 2.628284e+01 1.360221e+01 2.161929e+01 1.040322e+02
## IGFBP7 4.9647521 7.721845e+00 9.257950e+01 1.063388e+01 1.871761e+00
## IL1B 11.5082331 5.786362e+00 1.873428e+02 8.459227e+00 1.082648e+01
## IL6R 13.7341719 1.386817e+01 1.317601e+01 2.699827e+00 1.613570e+00
## IL7R 275.9560156 3.761636e+02 1.901201e+01 2.676964e+02 3.417850e+01
## IQGAP2 7.0274747 1.196006e+01 1.039990e+01 9.504364e+00 1.735927e+00
## IRF1 32.8746387 3.663196e+01 2.815950e+01 5.114372e+01 3.488356e+01
## ISG15 675.0958514 1.454041e+03 2.404475e+03 7.588962e+02 7.332002e+02
## ITM2C 30.9308319 3.142828e+01 3.246676e+00 2.773345e+01 1.398515e+02
## JAK1 20.0571216 3.221298e+01 9.766351e+00 2.609797e+01 2.062581e+01
## JUN 1066.5059079 1.364165e+03 4.192764e+02 1.042651e+03 9.868035e+02
## KDM1B 3.9106023 1.907179e+00 8.256585e+00 8.033592e-01 6.352560e+00
## KHDRBS1 97.7667917 1.471279e+02 7.368637e+01 1.132815e+02 6.968025e+01
## KLF3 21.6609279 1.623889e+01 1.567121e+01 1.453292e+01 1.967833e+01
## KYNU 0.4599142 4.880376e-01 7.913858e+00 4.377939e-01 5.802564e+00
## LAMTOR2 137.8637341 1.763239e+02 3.807696e+02 2.495174e+02 1.676277e+02
## LAMTOR5 115.1162704 1.450059e+02 2.092146e+02 1.689227e+02 1.936675e+02
## LAP3 31.5602392 3.596621e+01 8.308118e+01 4.404127e+01 4.527894e+01
## LAPTM4A 159.1533695 1.666000e+02 1.838326e+02 1.799602e+02 2.129680e+02
## LARP1 5.6805090 5.839856e+00 7.765324e+00 4.327487e+00 6.521173e+00
## LCK 303.5720642 3.206761e+02 1.959634e+01 3.651793e+02 4.045600e+01
## LDLRAP1 74.0789533 5.314029e+01 4.758809e+00 7.877455e+01 6.875446e+00
## LEF1 118.5815548 7.980122e+01 8.248355e+00 1.445161e+01 5.950043e+00
## LEPROT 14.2347569 1.199015e+01 2.338085e+01 1.308293e+01 1.080782e+01
## LGALSL 0.4455760 0.000000e+00 3.066115e-01 0.000000e+00 6.138174e-01
## LIMS1 26.2785169 4.221962e+01 1.793442e+01 1.502943e+01 9.622016e+00
## LINC00877 0.0000000 8.323552e-01 8.371159e+00 6.796460e-01 1.570522e+00
## LINC01011 4.8507991 4.763102e+00 2.105627e+00 3.187231e+00 1.220115e+00
## LMNA 4.0306246 2.586306e+01 5.985924e+00 9.659359e+00 2.752535e+00
## LPCAT1 5.9965002 4.335068e+00 6.893332e+00 1.594064e+01 8.335790e+00
## LRPAP1 14.1167008 2.025375e+01 1.981567e+01 2.378009e+01 1.683220e+01
## LRRFIP1 35.7309293 4.420025e+01 5.038628e+01 3.426575e+01 3.471370e+01
## LSM10 120.0386326 1.556902e+02 1.899830e+02 1.808568e+02 3.450699e+02
## LSM6 49.9311533 5.225334e+01 6.920685e+01 3.892541e+01 3.809251e+01
## LST1 81.5784231 1.193269e+02 2.154994e+03 8.607368e+01 9.321168e+01
## LTB 3868.8925289 6.190168e+03 2.089734e+02 2.551901e+03 4.975571e+03
## LY6G6F 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## LY86 18.6637378 2.597773e+01 6.321357e+02 1.076670e+01 8.490878e+02
## LYAR 65.6259321 1.819453e+02 7.035402e+01 5.818783e+02 3.468663e+01
## LYST 1.3768295 1.667912e+00 9.483296e+00 5.489734e+00 7.305134e+00
## MAD2L2 11.0607118 1.779998e+01 1.595938e+01 2.411117e+01 1.496627e+01
## MAGEF1 66.1461590 5.106555e+01 1.552577e+01 4.022799e+01 4.715376e+01
## MAL 381.3785944 3.249485e+02 1.184484e+01 2.969472e+01 2.239378e+01
## MANF 65.6595597 9.894512e+01 4.160711e+01 1.209084e+02 6.291721e+01
## MAPKAPK3 21.8991366 2.314955e+01 5.356168e+01 2.477516e+01 1.099074e+01
## MARCKSL1 162.1202020 1.219649e+02 1.857584e+02 9.626900e+01 3.927644e+02
## MARCO 0.5799827 1.984892e+00 8.509553e+01 0.000000e+00 0.000000e+00
## MBD4 47.2184930 2.225696e+01 2.021426e+01 3.137625e+01 9.729957e+01
## MBNL1-AS1 1.0106649 5.609651e-01 2.224902e+00 2.656104e+00 1.460317e+00
## MCL1 97.1851156 1.300549e+02 1.436949e+02 1.242763e+02 6.648094e+01
## MCUR1 8.7685910 1.092605e+01 6.636245e+00 8.275837e+00 1.425786e+01
## MEF2C 1.6250892 6.093019e-01 8.543663e+00 1.775946e+00 4.720823e+01
## MFSD1 8.7805690 6.628699e+00 2.398819e+01 7.544559e+00 1.401744e+01
## MGAT4A 29.3273973 2.776105e+01 2.093962e+00 2.521439e+01 6.683809e+00
## MGLL 0.3454315 2.015582e-01 2.724473e+00 6.424097e-01 0.000000e+00
## MGST2 5.0652359 7.197818e+00 3.421467e+01 5.851491e+00 1.314749e+00
## MIB2 8.2200833 9.168824e+00 1.739003e+00 2.691206e+01 6.131167e+00
## MNDA 9.3266984 1.245308e+01 3.031089e+02 9.202988e+00 3.858500e+01
## MOB1A 27.7904777 3.115077e+01 4.394515e+01 3.115055e+01 5.699219e+01
## MOB3C 2.6346030 4.968566e+00 2.760058e+00 7.283856e+00 1.721598e+00
## MPZL1 0.8383305 5.145556e-01 2.039247e+00 0.000000e+00 1.247238e+00
## MRPL53 157.5170900 1.188734e+02 1.560813e+02 1.195964e+02 2.044659e+02
## MTHFD2 4.8063734 6.442972e+00 9.490032e+00 3.591522e+00 3.666622e+00
## MTMR11 1.2435003 4.560315e-01 4.257577e+01 0.000000e+00 0.000000e+00
## MTMR14 15.7978710 1.671654e+01 2.817804e+01 1.792381e+01 2.733822e+01
## MXD3 1.8123788 3.403994e+00 5.509247e+00 0.000000e+00 1.007406e+00
## MXD4 3.8268848 4.385195e+00 6.802751e+00 4.336712e+00 5.395340e+00
## MYCL 2.0499061 0.000000e+00 1.557730e+01 0.000000e+00 0.000000e+00
## MYD88 22.8427231 3.630153e+01 5.004018e+01 2.899745e+01 1.917718e+01
## MYLK 0.0000000 4.352927e-02 0.000000e+00 1.664945e-01 0.000000e+00
## MZB1 4.8931201 2.832626e+00 2.115185e+00 6.078399e+00 1.255568e+02
## MZT2A 125.9994473 1.667744e+02 3.662695e+01 1.115294e+02 8.428731e+01
## NAAA 33.8299557 4.174111e+01 8.473448e+01 4.516225e+01 2.787908e+01
## NADK 1.6355905 4.334366e+00 1.661387e+01 7.220992e+00 5.757459e+00
## NAGK 8.2769406 1.147183e+01 3.677810e+01 1.095776e+01 8.250792e+00
## NBPF10 2.2972006 1.791871e+00 1.210596e+01 3.225556e+00 9.415889e+00
## NCF2 6.8907180 1.146484e+01 2.410224e+02 1.009860e+01 1.734311e+01
## NCR3 45.8848165 7.158958e+01 5.223642e+00 5.810485e+02 1.427396e+02
## NDFIP1 166.8117298 1.081821e+02 4.996578e+01 6.684987e+01 3.161415e+01
## NDUFA2 365.7444539 2.074423e+02 3.151872e+02 2.779938e+02 1.836998e+02
## NDUFB3 81.0235075 8.831083e+01 1.107421e+02 1.232479e+02 7.757872e+01
## NDUFB5 50.3743317 4.713635e+01 4.608707e+01 4.529665e+01 4.161635e+01
## NDUFS6 265.5592926 3.120096e+02 5.148473e+02 3.982123e+02 2.108531e+02
## NEXN 0.2575275 5.701809e-01 3.557068e+00 0.000000e+00 3.426991e+00
## NFKBIZ 32.1024772 2.101815e+01 3.401102e+01 2.277322e+01 1.422651e+01
## NMI 43.0765735 7.619672e+01 7.669752e+01 5.362458e+01 4.396513e+01
## NOTCH2 1.9103245 1.433792e+00 4.753020e+00 2.074700e+00 4.186724e+00
## NPL 1.3640540 3.372672e+00 2.277886e+01 3.338695e+00 2.314528e+00
## NPM1 575.4695672 4.568064e+02 1.442280e+02 3.525555e+02 3.721163e+02
## NQO2 29.7184820 3.373259e+01 6.985434e+01 3.870728e+01 1.314260e+01
## NREP 0.3468873 0.000000e+00 9.623065e-01 0.000000e+00 0.000000e+00
## NUDT16 2.7642331 4.941330e+00 3.307751e+01 7.295403e+00 2.736494e+00
## NUDT3 5.9739590 8.137305e+00 1.191774e+01 8.784323e+00 1.330079e+01
## NUP210 7.8066236 5.830577e+00 6.232072e+00 1.009243e+01 4.486293e+00
## OCIAD2 215.5570397 2.195865e+02 1.416009e+01 1.947578e+02 1.288719e+02
## ODC1 37.1708717 8.622176e+01 1.417271e+01 2.739048e+01 9.680278e+01
## ODF2L 17.1591332 2.214352e+01 2.022143e+00 2.587079e+01 1.092682e+01
## OST4 1947.8523171 2.136456e+03 1.196768e+03 2.125733e+03 1.506644e+03
## OXNAD1 56.9274264 3.499814e+01 5.660437e+00 3.085778e+01 8.270188e+00
## P2RY13 0.8611559 1.037177e+00 1.075923e+01 0.000000e+00 0.000000e+00
## PAFAH2 2.7897407 4.585552e-01 2.758426e+00 9.713069e+00 3.244130e+00
## PAIP1 18.3009581 2.631365e+01 1.975810e+01 1.961451e+01 1.840023e+01
## PAPSS1 11.0290824 1.596315e+01 9.466851e+00 1.118948e+01 2.245601e+01
## PARL 59.5745319 5.182919e+01 1.439012e+02 6.452823e+01 5.820629e+01
## PARP1 16.5860721 9.212640e+00 2.810454e+00 8.573293e+00 2.235115e+01
## PBX1 0.0000000 0.000000e+00 1.450768e-01 0.000000e+00 0.000000e+00
## PBXIP1 67.4691877 1.077480e+02 1.094665e+01 9.657931e+01 3.776575e+01
## PDCL3 111.7163375 1.172204e+02 5.368571e+01 1.067560e+02 4.581506e+01
## PDIA6 57.8274839 8.285557e+01 6.202551e+01 1.102731e+02 7.811362e+01
## PDLIM5 3.8903332 4.610537e+00 3.907021e+00 4.395529e+00 3.571539e+00
## PDZK1IP1 1.7224551 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## PF4 4.2174172 2.722630e+00 3.791010e+01 2.908335e+01 1.676028e+01
## PGD 22.1254961 1.966591e+01 1.231255e+02 1.626614e+01 2.245113e+01
## PHACTR1 0.2917842 1.060998e-01 5.794662e+00 9.526768e-02 8.203533e+00
## PID1 2.2833382 1.810830e+00 3.790816e+01 1.051905e+00 0.000000e+00
## PIM1 119.0624699 1.423889e+02 1.669023e+01 1.673491e+02 3.326474e+01
## PLA2G12A 13.0778360 2.344000e+01 5.291033e+00 8.367328e+00 8.512326e+00
## PLAC8 177.8747503 1.886565e+02 1.794158e+02 2.385479e+02 4.740713e+02
## PLEK 13.6573714 1.730890e+01 7.733914e+01 1.555836e+02 7.284092e+01
## PLEKHO1 5.8903052 8.694286e+00 2.653334e+01 4.357049e+00 1.459726e+01
## PLSCR1 18.0271469 2.896857e+01 7.131767e+01 3.669141e+01 3.050894e+01
## PMVK 143.4491660 2.160572e+02 2.118835e+02 1.622653e+02 1.910111e+02
## POLE4 85.8047616 9.610607e+01 2.250252e+02 7.618431e+01 1.175194e+02
## PPBP 12.7376732 1.401306e+01 7.343549e+01 6.340666e+01 2.157636e+01
## PPCS 64.7083705 7.882061e+01 6.480395e+01 5.666505e+01 8.548266e+01
## PPP1R18 59.9358834 9.233713e+01 6.704042e+01 1.625359e+02 7.520304e+01
## PPP3R1 21.4573470 1.629779e+01 1.227045e+01 2.271299e+01 2.647558e+01
## PPT1 65.1853033 7.547269e+01 1.370899e+02 4.437712e+01 4.301818e+01
## PRADC1 26.5645082 2.893180e+01 3.202918e+01 3.347954e+01 2.146726e+01
## PRDX1 231.3410800 3.688155e+02 2.837788e+02 2.798941e+02 2.544532e+02
## PRDX6 147.4728647 1.444954e+02 1.943191e+02 1.980748e+02 1.475219e+02
## PRELID1 223.9846835 3.295090e+02 9.261244e+02 4.252927e+02 2.953426e+02
## PRKCD 5.1971343 1.983206e+00 3.344955e+01 7.342309e+00 1.110671e+01
## PRPF40A 16.8840358 1.820784e+01 1.228360e+01 1.960275e+01 1.015437e+01
## PSMA5 47.7629421 7.889631e+01 4.973145e+01 8.242763e+01 4.924714e+01
## PSMB4 85.6471019 1.149595e+02 1.287304e+02 9.799684e+01 9.814774e+01
## PSMB8 152.5432505 2.867052e+02 2.100304e+02 2.981409e+02 2.628377e+02
## PSMB9 221.8630141 3.980229e+02 3.448616e+02 4.263375e+02 3.440603e+02
## PSRC1 0.0000000 2.086860e+00 3.776780e+00 0.000000e+00 0.000000e+00
## PTCRA 3.3277465 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## PTP4A2 34.0407393 4.601453e+01 4.583708e+01 7.548547e+01 4.336549e+01
## PTPN18 20.6178069 1.653954e+01 4.004622e+01 2.644079e+01 3.566720e+01
## PTPN4 8.6623839 1.059377e+01 7.001435e-01 1.769682e+01 5.419383e+00
## PTPN7 27.4356345 2.117558e+01 4.236268e+00 3.274379e+01 2.291482e+01
## PYHIN1 15.3074050 3.571751e+01 3.954079e+00 5.091098e+01 3.173854e+00
## QPCT 2.6120888 1.379892e+00 9.924286e+01 1.364206e+00 3.042440e+00
## RAB10 30.7743477 4.989144e+01 7.094442e+01 3.861207e+01 4.937099e+01
## RAB13 3.6293535 0.000000e+00 2.201884e+01 1.388278e+00 1.735627e+00
## RAB24 22.5079353 2.222689e+01 1.099639e+02 1.718806e+01 6.597847e+01
## RABGAP1L 4.1404382 7.413699e+00 7.844897e+00 5.758591e+00 5.917845e+00
## RAD50 6.9821058 6.435140e+00 3.357288e+00 7.347120e+00 5.990681e+00
## RALB 20.7963800 2.750234e+01 4.351557e+01 2.852004e+01 4.881621e+01
## RALGPS2 2.1340823 1.864296e+00 4.988005e-01 6.386421e-01 3.187323e+01
## RAMP1 2.4317458 3.061187e+00 4.453504e+00 2.175734e+01 0.000000e+00
## RASSF5 34.4030795 4.010101e+01 1.468104e+01 5.033607e+01 2.102479e+01
## RBP7 9.9741873 0.000000e+00 6.023719e+02 0.000000e+00 1.801837e+01
## RCAN3 40.2308939 3.464887e+01 5.006215e+00 1.050396e+01 1.381829e+01
## RCC2 3.7594813 4.722449e+00 1.705072e+01 8.807204e+00 4.767511e+00
## RCSD1 30.6847911 3.426014e+01 2.686783e+01 4.171366e+01 8.092648e+01
## REEP5 47.3362276 7.511768e+01 1.293755e+02 1.231039e+02 9.444379e+01
## REL 6.0302780 4.126972e+00 6.841647e+00 9.014959e+00 2.198515e+01
## RER1 31.2878278 7.598537e+01 5.707438e+01 6.119575e+01 6.725077e+01
## RETSAT 2.8762266 4.005801e+00 1.061604e+00 3.385289e+00 8.876910e-01
## RGS1 35.3914926 1.018661e+02 1.329997e+01 5.584910e+01 4.344866e+01
## RGS18 1.9429998 3.387723e+00 3.026261e+01 1.009618e+01 2.741534e+00
## RGS2 67.0864112 1.200328e+02 5.738760e+02 1.875122e+02 1.590351e+02
## RHOA 165.7973773 1.986759e+02 2.976125e+02 2.383269e+02 2.795685e+02
## RHOB 22.8467864 3.326591e+01 1.899537e+02 3.567664e+01 4.354488e+01
## RHOC 15.7976539 3.724998e+01 4.807001e+01 7.692056e+01 3.458491e+01
## RHOH 108.7665834 8.514428e+01 3.084679e+00 5.923572e+01 9.431179e+01
## RNF11 32.0635773 3.081236e+01 1.701717e+01 1.919355e+01 3.213729e+00
## RNF13 22.4059381 2.968245e+01 3.473690e+01 3.259639e+01 3.271132e+01
## RNF130 4.7660920 4.830964e+00 6.241682e+01 5.752666e+00 6.923685e+00
## RNF144B 1.9067359 1.746923e+00 5.789295e+00 1.241515e+00 1.337060e+00
## RNF149 25.2649843 3.667244e+01 3.745432e+01 4.126396e+01 1.678429e+01
## RNF181 170.2348448 1.741600e+02 2.999582e+02 4.227883e+02 1.124840e+02
## RNF7 124.9073291 9.810348e+01 1.440068e+02 9.684722e+01 5.957849e+01
## RNPEP 21.0451689 1.452927e+01 5.361847e+01 1.487183e+01 2.407861e+01
## RPA2 164.7858853 1.545596e+02 3.065910e+01 2.030816e+02 1.217803e+02
## RPL22 812.5714723 6.153136e+02 3.157844e+02 4.468313e+02 6.179345e+02
## RPL22L1 113.8705283 1.311419e+02 6.305233e+01 1.449759e+02 3.632936e+02
## RRAGC 10.2514350 1.484129e+01 1.032141e+01 6.072161e+01 1.409163e+01
## RSAD2 9.9148252 1.505432e+01 1.805938e+01 6.399531e+00 5.764682e+00
## RSPH9 0.0000000 0.000000e+00 2.302756e+00 0.000000e+00 0.000000e+00
## RTN4 32.0011556 5.204376e+01 5.605187e+01 6.041354e+01 3.737396e+01
## RUFY1 10.7591613 8.522214e+00 1.248348e+01 1.244356e+01 1.196924e+01
## RUNX3 15.9070510 1.579117e+01 1.207397e+01 4.815040e+01 1.313948e+01
## S100A10 1082.6823611 2.387398e+03 3.179371e+03 1.843457e+03 4.789002e+02
## S100A11 740.7410634 2.042693e+03 6.659858e+03 1.474188e+03 3.292014e+02
## S100A12 9.0483988 0.000000e+00 1.190273e+03 6.056188e+00 1.780866e+01
## S100A4 2314.3072515 7.639561e+03 1.565684e+04 7.545587e+03 8.804761e+02
## S100A6 1828.1084634 2.918918e+03 1.038267e+04 3.820459e+03 7.883320e+02
## S100A8 144.0159042 1.495510e+02 1.904883e+04 5.970299e+01 1.264505e+02
## S100A9 375.5251297 3.625921e+02 5.030501e+04 4.029636e+02 4.332952e+02
## SCARB2 0.9365993 3.128667e-01 2.734933e+00 6.016769e-01 6.282259e-01
## SCFD2 1.8125961 4.829026e+00 2.333383e+00 3.274133e+00 2.521623e+00
## SCP2 32.0318301 5.595819e+01 3.465724e+01 5.366351e+01 4.703379e+01
## SCYL3 3.9980059 4.889326e+00 2.704018e+00 5.330989e+00 3.099656e+00
## SDF4 29.1651399 4.467715e+01 2.189565e+01 3.643185e+01 2.124982e+01
## SDHB 70.2500388 1.311938e+02 1.970994e+02 6.247441e+01 9.750402e+01
## SEC24D 0.2174049 5.433767e-01 8.902124e-01 2.215160e+00 4.139326e-01
## SEC62 46.6530115 3.797462e+01 1.814220e+01 3.950739e+01 8.806650e+01
## SEL1L3 2.6352994 5.337440e+00 1.642565e+00 5.460338e+00 3.357656e+01
## SELL 553.5352402 4.066767e+02 1.494268e+02 2.660586e+02 7.453111e+02
## SEMA4A 1.6978882 2.125724e+00 1.212402e+01 1.515152e+00 6.914926e+00
## SERP1 186.9051257 1.720074e+02 2.435546e+02 1.685443e+02 1.668123e+02
## SERPINB1 152.7648435 1.367944e+02 3.952351e+02 1.448687e+02 1.045189e+02
## SERPINB6 13.5341800 1.446608e+01 2.926522e+01 1.574934e+01 1.212258e+01
## SERPINE2 2.4314479 1.938462e-01 1.411067e-01 5.200925e-01 5.201774e-01
## SF3B4 32.6284838 5.797257e+01 3.140099e+01 9.713593e+01 6.061719e+01
## SH2D1B 1.6042755 8.497654e-01 1.830648e+00 6.844947e+00 1.088870e+00
## SH2D2A 18.5690403 3.802328e+01 6.670932e+00 9.333024e+01 5.309240e+00
## SH3BGRL3 2346.4059408 4.383180e+03 6.448130e+03 5.407373e+03 2.565991e+03
## SH3BP2 3.1568247 2.595010e+00 9.244039e+00 5.078045e+00 4.480755e+00
## SH3BP5 24.5437981 6.158485e+01 1.171368e+01 4.888120e+01 4.556993e+01
## SH3D19 4.3945343 3.544339e+00 1.273166e+00 1.624014e+00 2.267500e+00
## SH3GLB1 20.3767384 2.459062e+01 3.184823e+01 5.285852e+01 2.167100e+01
## SH3RF1 0.4385763 0.000000e+00 1.294832e+00 0.000000e+00 1.849426e+00
## SH3YL1 26.2074780 1.994525e+01 1.086391e+00 8.697622e+00 7.308988e+00
## SKP1 68.3422913 8.570893e+01 2.976756e+01 7.203690e+01 5.805031e+01
## SLAMF7 1.5796345 9.730699e-01 2.859933e+00 2.044470e+01 0.000000e+00
## SLC11A1 1.5662443 2.696880e+00 3.802085e+01 1.305431e+00 6.128446e+00
## SLC30A9 2.6951336 4.310186e+00 3.854921e+00 2.814009e+00 2.826007e+00
## SLC39A1 11.0533446 3.772812e+01 2.056228e+01 1.974764e+01 1.591036e+01
## SLC40A1 19.4492102 1.517933e+01 5.503623e+00 3.227887e+00 2.531703e+00
## SMAP2 52.2597631 4.221943e+01 6.840679e+01 4.043449e+01 1.407557e+02
## SMIM14 6.3923062 8.864262e+00 1.447423e+01 1.512154e+01 1.169967e+02
## SNAPIN 59.1490526 8.287629e+01 1.533394e+02 1.211119e+02 1.077238e+02
## SNCA 0.7459595 0.000000e+00 9.599304e+00 1.601591e+00 0.000000e+00
## SNX18 3.2275977 2.347585e+00 5.133258e+00 3.492361e+00 5.462450e+00
## SNX2 17.2870709 2.137056e+01 3.247087e+01 1.970889e+01 1.168277e+02
## SNX27 1.6488021 1.120696e+00 5.613469e+00 1.953878e+00 2.641272e+00
## SP110 35.4898632 3.428370e+01 3.602394e+01 2.958236e+01 6.383571e+01
## SPARC 1.5064230 2.009954e+00 4.150436e+00 3.147718e+00 3.213186e+00
## SPATS2L 1.2818599 6.485933e+00 3.139124e+00 4.959880e+00 2.786383e-01
## SPOCD1 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## SPON2 6.5222137 5.352111e+00 3.677366e+00 4.321814e+01 3.232755e+00
## SRA1 22.8473241 2.955796e+01 1.198933e+02 3.916788e+01 4.162737e+01
## SRD5A3 4.4263246 3.962293e+00 9.841730e+00 7.055149e+00 6.356880e+00
## SSR3 30.1191532 4.722691e+01 6.581771e+01 4.943120e+01 3.517676e+01
## SSX2IP 0.4597245 4.871054e-01 0.000000e+00 5.198128e-01 0.000000e+00
## ST3GAL3 0.5800250 8.388598e-01 1.250538e-01 4.844137e-01 8.229658e-01
## ST3GAL5 2.6073153 2.465124e+00 3.317520e+00 3.661694e+00 2.385291e+00
## ST3GAL6 0.0000000 3.680392e-01 3.219502e+00 0.000000e+00 0.000000e+00
## STAT4 14.1651264 1.348624e+01 1.321765e+00 2.351464e+01 2.731939e+00
## STMN1 20.5334332 1.178493e+01 2.175296e+00 3.209304e+01 2.833717e+01
## STX18 5.6095259 1.243056e+01 6.019594e+00 5.495065e+01 1.230125e+01
## SYTL1 32.5636620 5.345399e+01 1.321548e+01 5.540921e+01 3.664659e+01
## TADA3 23.3329456 3.724227e+01 2.606542e+01 4.348997e+01 2.666621e+01
## TAGLN2 332.5269763 4.516703e+02 3.998366e+02 5.404950e+02 5.538699e+02
## TANK 20.5285321 3.280050e+01 2.099105e+01 1.832289e+01 3.972460e+01
## TAP1 55.1052025 9.742501e+01 3.431784e+01 1.066581e+02 6.039981e+01
## TARBP1 11.1198242 7.663431e+00 2.086278e+00 5.589800e+00 3.156280e+00
## TBC1D9 0.2601235 5.807599e-01 2.613819e+00 0.000000e+00 6.514698e+00
## TCF7 66.5777563 6.003042e+01 9.265573e+00 2.969622e+01 1.474735e+01
## TFDP2 3.3836562 4.702797e+00 1.387816e+00 6.159228e+00 6.842673e+00
## TGFBI 3.4767694 2.300135e+00 5.470713e+01 4.628517e+00 2.288229e+00
## THEMIS2 9.0077040 1.622170e+01 7.048126e+01 1.576706e+01 6.395146e+00
## TIGIT 24.3115427 2.875735e+01 1.289226e+00 6.786530e+01 9.910823e-01
## TKT 25.5694664 3.295417e+01 1.885250e+02 3.710993e+01 4.263158e+01
## TLR10 0.0000000 1.044696e+00 2.789883e+00 5.531308e-01 4.945266e+01
## TMCC2 0.0000000 0.000000e+00 0.000000e+00 8.861686e-01 1.552869e+00
## TMED9 93.3239218 1.188901e+02 5.401873e+01 1.627824e+02 1.072963e+02
## TMEM154 4.9660935 5.914035e+00 6.843713e+00 3.857822e+00 2.482167e+01
## TMEM40 0.0000000 0.000000e+00 5.723581e-01 1.295389e+00 0.000000e+00
## TMEM50A 102.7358522 1.313320e+02 8.719937e+01 2.992079e+02 7.781798e+01
## TMEM69 38.7138921 3.161944e+01 4.226590e+01 3.289704e+01 5.884871e+01
## TNFAIP8 51.4351785 8.697868e+01 1.238435e+01 4.335295e+01 5.831756e+01
## TNFRSF14 45.8190034 5.293868e+01 3.124011e+01 5.101452e+01 5.449154e+01
## TNFRSF1B 19.0610691 3.487578e+01 1.105002e+02 3.065416e+01 1.896616e+01
## TNFRSF25 23.0598746 6.503333e+01 2.033836e+00 2.583711e+01 3.283092e-01
## TNFRSF4 25.2816633 1.748703e+02 1.971065e+00 1.909795e+01 5.952652e+00
## TNFRSF8 0.3949028 4.206825e-01 5.551313e+00 0.000000e+00 0.000000e+00
## TNFSF10 55.8390912 1.017839e+02 1.724369e+02 4.887889e+01 5.043588e+01
## TNFSF4 0.4805866 6.913379e-01 0.000000e+00 0.000000e+00 1.470702e+00
## TNS1 0.0000000 1.821967e-01 4.163794e-01 0.000000e+00 0.000000e+00
## TPM3 42.4323988 5.442785e+01 4.429448e+01 5.359000e+01 5.596034e+01
## TPMT 13.8463902 1.945732e+01 3.374957e+01 2.522125e+01 4.638034e+00
## TRABD2A 35.5881120 3.058271e+01 2.286907e+00 5.352708e+00 6.855627e+00
## TRAF3IP3 187.6356585 1.532644e+02 4.078238e+01 1.514046e+02 1.206675e+02
## TRAK1 1.9758788 4.518122e-01 2.246777e+00 8.456039e-01 2.435914e+00
## TRAPPC12 5.0913302 5.259181e+00 6.532708e+00 5.664553e+00 1.089837e+01
## TRAPPC13 3.6743325 4.484614e+00 3.275052e+00 2.974543e+00 1.290539e+00
## TRAT1 175.2250076 2.257544e+02 7.439176e+00 1.167080e+02 1.408186e+01
## TREM1 0.3914530 1.734478e+00 2.484473e+01 2.395373e-01 5.300576e+00
## TREML1 0.0000000 1.310177e+00 2.792629e+00 0.000000e+00 5.903793e+00
## TREX1 49.9373358 1.304408e+02 1.755417e+02 8.589738e+01 1.174857e+02
## TRIM33 5.7499607 3.936051e+00 1.691558e+00 3.031691e+00 4.632154e+00
## TRIM58 0.8234670 0.000000e+00 0.000000e+00 1.254932e+00 0.000000e+00
## TSTD1 336.7490467 4.082086e+02 6.351356e+01 2.826581e+02 4.853622e+02
## TUBA4A 103.8557337 1.567383e+02 4.216380e+01 1.636744e+02 7.513259e+01
## TUBB2A 6.0297009 1.246275e+01 6.912448e+00 1.054584e+01 4.910471e+00
## TXK 60.4328485 3.980973e+01 4.234475e+00 2.587259e+01 3.702758e+00
## TXNIP 609.1638331 6.356599e+02 3.044366e+02 8.227370e+02 8.303427e+02
## UBE2B 70.5672893 5.542735e+01 3.717390e+01 5.732849e+01 4.852586e+01
## UBE2E2 3.4434525 1.070550e+00 2.204172e+01 1.283691e+00 4.248656e+01
## UBE2F 43.1643176 2.075003e+01 4.096002e+01 3.490884e+01 2.540902e+01
## UBXN11 10.3084678 1.754341e+01 4.011823e+01 1.372492e+01 1.005223e+01
## UNC50 35.9476305 3.282535e+01 2.854639e+01 5.156638e+01 4.104925e+01
## UQCRC1 25.7640213 2.559280e+01 5.905858e+01 3.255929e+01 1.966043e+01
## USP33 11.6806861 8.602999e+00 3.093148e+00 9.258051e+00 1.439805e+01
## VAMP3 17.5790516 2.587785e+01 4.602359e+01 2.042622e+01 1.909681e+01
## VAMP5 262.7199366 5.268200e+02 6.847972e+02 4.085685e+02 2.061760e+02
## VAMP8 944.0470426 1.145343e+03 1.490971e+03 1.021402e+03 5.849690e+02
## VCAN 1.8828359 2.183994e+00 3.299211e+01 3.634221e+00 2.525004e+00
## VDAC1 69.1925252 9.444353e+01 8.931489e+01 8.918704e+01 8.264959e+01
## VIL1 0.9107756 0.000000e+00 0.000000e+00 0.000000e+00 6.985612e-01
## WASF2 34.4759016 4.154718e+01 5.432626e+01 4.177501e+01 5.093034e+01
## WDR1 17.9249566 2.230986e+01 1.477396e+01 2.304438e+01 1.000735e+01
## WDR41 3.2414943 2.486051e+00 7.574653e+00 5.152707e+00 6.439659e+00
## WIPF1 18.1386698 2.124383e+01 8.104462e+00 2.805344e+01 1.682601e+01
## XCL1 1.7403208 5.986814e+00 1.646623e+00 1.297073e+02 2.857708e+00
## XCL2 7.0429227 3.984829e+01 5.800528e+00 4.135063e+02 6.239159e+00
## YBX1 577.0140451 6.933709e+02 9.031958e+02 6.911464e+02 9.375963e+02
## YWHAQ 109.3841294 2.473490e+02 4.760038e+01 2.817920e+02 1.359401e+02
## ZAP70 41.6123883 4.629708e+01 6.216552e+00 9.240072e+01 4.686951e+00
## ZCCHC17 53.4682254 4.542062e+01 3.507229e+01 4.640730e+01 4.262405e+01
## ZEB2 1.9232818 2.159594e+00 5.453262e+00 3.442013e+00 4.708432e+00
## ZFP36L2 614.6833929 7.052350e+02 2.928070e+02 6.541620e+02 2.467445e+02
## ZNF593 82.3532873 1.392588e+02 1.371652e+02 1.002783e+02 6.009924e+01
## ABHD11 9.9484940 1.305146e+01 9.793885e+00 1.458638e+01 6.070055e+00
## ABI1 32.7002870 4.174356e+01 1.825179e+01 5.490728e+01 5.281047e+01
## ABRACL 466.4256808 4.752321e+02 2.265745e+02 4.085420e+02 2.753783e+02
## ACAT2 24.2263881 4.365241e+01 1.306828e+01 3.224454e+01 6.527493e+01
## ACOT9 3.8275689 1.127723e+01 8.779981e+00 1.023517e+01 1.059383e+01
## ACRBP 3.3370997 1.308112e+00 4.750743e+00 0.000000e+00 4.513703e+00
## ACTR1A 19.2321941 2.023798e+01 2.531113e+01 2.590466e+01 1.162181e+01
## ADAM28 1.1468368 1.305470e+00 9.855022e-01 9.820723e-01 3.631922e+01
## ADD3 57.4637617 5.654119e+01 1.059254e+01 4.814781e+01 3.999819e+01
## ADK 30.0358362 3.022818e+01 1.863646e+01 1.442765e+01 9.988521e+01
## ADO 5.7550229 4.789261e+00 3.600775e-01 7.200832e+00 4.007290e+00
## AGPAT2 18.3158653 2.391169e+01 8.077900e+01 2.602177e+01 3.896755e+01
## AHNAK 12.6084762 2.209542e+01 2.658636e+01 3.466363e+01 1.347638e+01
## AKIRIN2 33.4125324 4.659103e+01 8.320981e+01 7.924655e+01 8.928928e+01
## AKR1C3 4.7641876 2.885879e+00 0.000000e+00 1.928103e+01 0.000000e+00
## ALDH2 1.9932178 1.192342e+00 8.147796e+01 2.139379e+00 5.207572e+00
## ALDH3B1 0.8782585 2.744506e-01 1.095577e+01 1.810132e+00 5.999959e-01
## ALOX5 8.3336712 9.406682e+00 4.716715e+01 8.798849e+00 7.032580e+01
## ALOX5AP 143.5718910 3.623849e+02 9.909748e+01 3.238755e+02 2.725337e+02
## ANK1 0.0000000 7.802495e-01 0.000000e+00 0.000000e+00 2.516746e+00
## ANXA1 250.5715946 5.766186e+02 3.565759e+02 5.524103e+02 4.820239e+01
## AOAH 20.3051331 1.464475e+01 6.864486e+01 5.142608e+01 1.622826e+01
## AP1S2 34.2693075 4.017464e+01 3.477091e+02 2.903078e+01 4.591399e+01
## APIP 16.6177807 2.172968e+01 1.133551e+01 1.377169e+01 2.422002e+01
## APLP2 8.0222948 1.095659e+01 7.594263e+01 1.387194e+01 4.915806e+01
## AQP3 151.2168961 2.933381e+02 3.105355e+00 9.077167e+01 1.997039e+01
## ARAP1 3.1085998 3.098474e+00 1.367359e+01 1.668176e+00 5.850667e+00
## ARF3 16.3119455 2.011691e+01 2.022478e+01 2.439658e+01 1.899704e+01
## ARF5 169.9716040 1.964687e+02 2.986265e+02 2.239049e+02 2.111844e+02
## ARHGAP21 1.6900459 1.251805e+00 4.740509e-01 2.536970e-01 1.897732e-01
## ARHGAP4 23.0969236 3.312977e+01 2.811524e+01 2.185267e+01 3.175458e+01
## ARHGAP6 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 1.087305e+00
## ARL6IP4 330.6143122 3.201511e+02 3.438227e+02 3.232247e+02 2.362022e+02
## ARMC10 13.8179913 1.943860e+01 1.097352e+01 1.669201e+01 1.958982e+01
## ARPC1A 59.2368781 4.056444e+01 9.915335e+01 5.916737e+01 7.808611e+01
## ARPC1B 226.4962704 3.678145e+02 6.920991e+02 3.299841e+02 3.297930e+02
## ARPC3 526.0750332 6.429257e+02 1.301118e+03 7.045675e+02 1.183425e+03
## ARPC5L 46.8565010 6.612631e+01 2.437760e+01 1.542687e+02 6.710034e+01
## ARRB1 1.1380529 1.721765e+00 8.216958e+00 2.779061e+00 4.250223e-01
## ARRDC1 30.7133291 3.519335e+01 3.005802e+01 3.649163e+01 2.198000e+01
## ASAH1 10.9277973 1.227959e+01 5.165740e+01 9.945525e+00 1.758661e+01
## ASCL2 4.6112108 7.622165e+00 6.732709e+01 1.151305e+02 7.464267e+00
## ASL 1.6867749 5.169432e+00 7.780601e+00 7.635230e+00 5.965435e+00
## ATG16L2 4.7741472 4.902038e+00 1.616612e+01 6.167892e+00 6.762805e+00
## ATP2B1 6.7007561 1.469635e+01 1.095209e+01 1.132834e+01 1.974493e+01
## ATP6AP2 35.4483648 3.374341e+01 3.372918e+01 3.241973e+01 2.285333e+01
## ATP6V1F 521.2531307 5.632770e+02 1.490604e+03 5.821214e+02 6.119402e+02
## ATXN2 2.7706196 3.024193e+00 1.233440e+00 1.677770e+00 2.597732e+00
## BAG1 22.7982125 2.955659e+01 4.137846e+01 2.948361e+01 3.486805e+01
## BCAP29 7.4596313 7.367867e+00 4.770946e+00 1.183205e+01 9.376169e+00
## BCL7A 0.5847741 1.588753e+00 1.729777e-01 5.201151e-01 3.093948e+01
## BEX2 233.6801672 1.862111e+02 7.687019e+00 1.142236e+02 1.270475e+02
## BIN2 134.0589639 1.659303e+02 7.736484e+01 2.929363e+02 1.053471e+02
## BIRC3 35.5679121 4.415559e+01 5.106406e+00 1.333323e+01 7.185299e+01
## BLK 1.2334789 3.737856e+00 3.143529e-01 1.841763e+01 1.187043e+02
## BLNK 1.0384543 7.663238e-01 7.017152e-01 3.359807e-01 4.594621e+01
## BLOC1S1 230.1675735 2.189030e+02 3.625197e+02 3.857675e+02 2.640640e+02
## BLOC1S2 52.5457119 6.306049e+01 5.868031e+01 6.422172e+01 1.727097e+02
## BLVRA 46.4080190 9.210610e+01 4.394948e+02 5.890635e+01 3.697937e+01
## BMS1 3.8175779 4.605521e+00 4.629486e+00 6.312395e+00 1.446983e+01
## BNIP3L 48.9198834 3.330754e+01 1.062825e+02 2.977149e+01 4.531500e+01
## BPGM 15.3898208 2.500566e+01 6.510055e+00 3.172641e+01 1.363036e+01
## BRI3 17.0825104 1.634568e+01 1.847848e+02 9.733852e+00 9.301308e+00
## BRI3BP 2.0257006 2.299372e+00 4.534386e+00 2.521186e+00 7.031702e+00
## BTK 2.7181189 1.338680e+00 1.590898e+01 1.165792e+00 5.681660e+01
## BZW2 35.8176185 2.902907e+01 1.736032e+01 2.152941e+01 2.530777e+01
## C11orf21 14.2194647 7.103308e+00 2.771086e+01 1.945337e+01 1.038026e+01
## C11orf54 2.4198489 2.081069e+00 1.154151e+00 2.673087e+00 3.602785e+00
## C12orf57 509.4177427 3.508703e+02 7.898917e+01 3.614565e+02 1.883145e+02
## C12orf75 31.8805764 8.813823e+01 6.343369e+00 2.183850e+02 6.430383e+01
## C3AR1 2.6340983 2.631662e+00 1.110332e+01 9.361923e+00 1.178956e+00
## C7orf50 31.8589448 3.487757e+01 4.513807e+01 4.641282e+01 4.930269e+01
## C9orf78 101.4510403 1.098413e+02 4.063509e+01 1.042035e+02 6.964464e+01
## CA2 3.0310932 1.036431e+00 1.388536e+01 1.160727e+01 7.030382e+00
## CALHM2 9.3209817 2.043176e+01 4.289161e+01 3.831573e+01 9.373982e+00
## CAPN1 15.7011570 1.923242e+01 1.898200e+01 2.068113e+01 2.216942e+01
## CAPZA2 24.4394052 2.888736e+01 3.782823e+01 3.572507e+01 2.880664e+01
## CARD16 101.5201378 2.039075e+02 4.743082e+02 2.364332e+02 1.284635e+02
## CARD9 0.3358240 3.213429e-01 1.006230e+01 2.433563e+00 0.000000e+00
## CASP1 52.6321314 8.360407e+01 1.586819e+02 1.048286e+02 4.430548e+01
## CASP4 47.5697813 6.838501e+01 1.153997e+02 7.586946e+01 6.021450e+01
## CAT 38.9295090 3.903452e+01 6.747149e+01 4.010494e+01 7.419960e+01
## CATSPER1 0.6505484 1.371231e+00 5.713153e+01 0.000000e+00 0.000000e+00
## CCDC107 133.2690549 2.422408e+02 7.569974e+01 2.646821e+02 2.260225e+02
## CCDC22 20.1831959 4.244716e+01 1.926019e+01 1.243860e+01 2.258146e+01
## CCDC86 4.3787780 3.690913e+00 3.242911e+00 3.317512e+00 4.865073e+00
## CCDC90B 16.3601498 2.175276e+01 9.152174e+00 2.943521e+01 2.158649e+01
## CD151 2.3276355 1.036247e+01 2.278628e+01 1.213854e+01 5.703661e+00
## CD164 75.2729248 9.325942e+01 3.621967e+01 7.665581e+01 9.209561e+01
## CD27 538.0565332 5.254438e+02 1.991510e+01 4.936929e+02 1.547944e+02
## CD27-AS1 2.7782035 5.787258e+00 8.260856e+00 3.553060e+00 1.013351e+01
## CD36 0.5971051 1.626963e+00 2.344316e+01 9.962216e-01 7.175604e-01
## CD3D 915.3977164 9.143920e+02 3.103404e+01 1.077512e+03 2.546860e+01
## CD3E 790.3522759 8.212328e+02 3.661372e+01 8.049217e+02 6.050022e+01
## CD3G 146.4530127 1.590298e+02 6.639497e+00 2.011817e+02 1.697345e+01
## CD4 23.7256759 3.857970e+01 6.797786e+01 8.576812e+00 3.686426e+00
## CD40LG 33.9319548 5.905803e+01 3.020285e+00 1.492225e+01 4.553525e+00
## CD5 55.3439714 6.401890e+01 2.255294e+00 2.598063e+01 9.172635e+00
## CD6 43.2014008 4.376704e+01 3.480967e+00 3.432440e+01 5.374421e+00
## CD63 99.6352496 1.853264e+02 4.165999e+02 3.528999e+02 9.697649e+01
## CD69 347.4234663 4.499330e+02 3.006423e+01 2.917427e+02 4.757820e+02
## CD72 4.5024059 3.112899e+00 1.941057e+00 5.890136e+00 2.056296e+02
## CD82 20.9247931 3.570801e+01 5.074113e+00 1.062003e+01 6.381397e+01
## CD9 2.9691396 1.664301e+00 1.323690e+01 4.756164e+00 8.483471e+00
## CD99 142.1765222 2.660262e+02 1.781920e+02 3.835947e+02 1.006045e+02
## CDCA7L 15.5979361 2.840794e+00 2.819420e+00 1.914334e+00 6.761339e+01
## CDH23 0.5832937 2.098785e-01 2.018892e+00 2.017428e-01 6.621122e-01
## CDK2AP1 0.3127173 0.000000e+00 4.342431e+00 0.000000e+00 6.891431e-01
## CDKN1B 17.8363148 2.775110e+01 7.873163e+00 1.998409e+01 1.721423e+01
## CDKN1C 0.8144038 4.447993e+00 1.160380e+01 0.000000e+00 2.988657e+00
## CEBPD 44.5372330 4.830395e+01 1.176998e+03 2.403788e+02 6.291833e+01
## CFP 12.0461048 1.456066e+01 2.340977e+02 5.772535e+00 1.067794e+01
## CHCHD2 1363.9138837 1.612994e+03 1.452193e+03 1.715961e+03 1.541440e+03
## CHST12 4.6747579 1.018954e+01 1.653308e+00 2.610253e+01 6.631838e+00
## CHST7 2.2569253 1.041278e+01 8.022068e+00 5.378330e+00 2.382693e+00
## CKAP4 6.0172546 4.656550e+00 2.031688e+01 6.373421e-01 8.075358e+00
## CLEC12A 0.0000000 6.268662e-01 4.782119e+01 2.056193e+00 1.493877e+00
## CLEC1B 0.4557056 0.000000e+00 1.003546e+00 6.494259e-01 0.000000e+00
## CLEC4A 3.6806151 1.606391e+00 1.019661e+02 0.000000e+00 1.003713e+01
## CLEC4C 0.0000000 0.000000e+00 1.698390e+00 0.000000e+00 0.000000e+00
## CLEC4E 0.0000000 0.000000e+00 7.295898e+01 0.000000e+00 0.000000e+00
## CLEC7A 3.2335155 2.091742e+00 5.688391e+01 1.063277e+00 5.800159e+00
## CLIC2 0.8926756 0.000000e+00 1.769025e+00 0.000000e+00 0.000000e+00
## CLIC3 26.4941171 2.553368e+01 5.053860e+00 2.489028e+02 2.438292e+01
## CLN8 4.3960559 2.507182e+00 3.265714e+00 8.458741e-01 1.245335e+01
## CLTA 120.0193356 1.243544e+02 2.874630e+02 1.142795e+02 1.403636e+02
## CLU 1.7183074 1.718607e+00 2.385585e+00 5.910406e+00 4.454517e-01
## COMMD6 579.5259722 4.912602e+02 2.320387e+02 4.417226e+02 4.995628e+02
## COMMD9 17.1853956 1.643699e+01 3.317251e+01 1.518118e+01 2.559538e+01
## CORO1B 51.6706853 9.667283e+01 5.195373e+01 2.115101e+01 6.041388e+00
## COX6A1 502.2109514 6.326546e+02 6.050389e+02 6.109751e+02 6.690206e+02
## COX6C 350.3198301 3.659843e+02 1.459797e+02 3.057168e+02 1.824613e+02
## COX8A 823.3121713 1.131194e+03 1.644039e+03 1.151734e+03 8.114497e+02
## CPSF6 15.9733048 1.037273e+01 4.590689e+00 1.374298e+01 1.739029e+01
## CPVL 2.6864332 7.462284e+00 1.815967e+02 7.352837e+00 7.205174e+00
## CSF2RA 0.8571506 1.081641e+00 3.145952e+01 1.096077e+00 1.566992e+00
## CTNNAL1 0.6687368 1.610125e+00 0.000000e+00 4.035763e+00 1.556133e+00
## CTSB 10.2267144 1.337465e+01 8.293920e+01 1.096243e+01 8.294686e+00
## CTSC 6.4405593 1.313945e+01 1.350002e+01 3.041936e+01 4.812506e+00
## CTSD 32.5251906 2.929070e+01 1.468565e+02 5.504834e+01 1.298109e+01
## CTSL 5.5665296 5.644006e+00 1.136687e+01 2.950599e+00 9.222180e-01
## CTSW 239.9009432 2.373816e+02 3.237163e+01 1.770897e+03 5.832561e+01
## CTTN 0.0000000 0.000000e+00 9.815373e-01 0.000000e+00 0.000000e+00
## CUX1 3.7117779 1.192034e+00 4.567202e+00 3.118687e+00 3.593862e+00
## CYB561A3 6.0683805 5.949759e+00 6.741383e+00 6.437449e+00 6.142158e+01
## CYBB 4.4599442 7.085663e+00 7.460562e+01 5.280082e+00 3.560753e+01
## CYC1 87.9333213 9.046080e+01 1.637680e+02 1.268345e+02 1.721175e+02
## DBNL 18.1435632 2.042017e+01 2.348943e+01 2.756873e+01 3.398767e+01
## DCTN3 71.2089964 9.210175e+01 8.108095e+01 7.357998e+01 6.248952e+01
## DDIT4 199.9730486 2.530171e+02 6.905869e+01 2.091121e+02 1.472428e+02
## DENND1A 1.6081542 1.455638e+00 6.484331e+00 3.525449e-01 1.909543e+00
## DERL1 14.9062919 4.511751e+01 1.262104e+01 2.835860e+01 1.548195e+01
## DMTN 1.1464467 3.889618e+00 0.000000e+00 3.868894e+00 5.178127e-01
## DNAJB6 14.4155175 1.791150e+01 1.220046e+01 1.964369e+01 8.725917e+00
## DNAJB9 94.4334571 1.306147e+02 1.941105e+01 7.368433e+01 1.192673e+02
## DNAJC15 19.3012585 3.396852e+01 1.921546e+01 2.163645e+01 2.987970e+01
## DNAJC4 44.6930984 3.820039e+01 7.266062e+01 3.091507e+01 9.212665e+01
## DOK2 69.5140362 2.526266e+02 2.658738e+02 2.488420e+02 2.021363e+01
## DPYSL2 0.5698980 2.469196e+00 7.013730e+00 1.141415e+00 0.000000e+00
## DRAP1 432.2888142 3.808027e+02 3.907423e+02 5.065603e+02 5.195952e+02
## DUSP5 10.9304044 1.737409e+01 1.047527e+01 1.122001e+01 1.696940e+01
## DUSP6 9.4040683 1.273881e+01 7.399699e+01 5.428562e+00 1.394237e+01
## DYNLL1 108.0568735 1.808128e+02 2.236438e+02 1.718507e+02 1.553146e+02
## DYNLT1 51.3509431 1.129467e+02 1.728935e+02 8.483342e+01 3.261607e+01
## EBP 62.0290832 1.060234e+02 3.710718e+01 1.267717e+02 6.460043e+01
## EFNB1 0.0000000 2.379193e+00 3.743113e+00 2.320787e+00 0.000000e+00
## EGFL7 4.8368981 0.000000e+00 5.505672e-01 0.000000e+00 0.000000e+00
## EHBP1L1 2.4680420 3.153535e+00 1.584363e+01 5.702138e+00 4.058971e+00
## EI24 31.7507253 2.466062e+01 1.755947e+01 3.005562e+01 1.809958e+01
## EIF4EBP1 52.6134433 4.874295e+01 4.534884e+02 6.696726e+01 1.354080e+02
## ENDOD1 0.9174163 2.187062e+00 1.773826e+00 3.973272e+00 6.339841e+00
## ENDOG 15.0020494 9.421846e+00 2.143445e+01 1.386011e+01 1.842452e+00
## ENHO 2.4926122 4.541801e+00 1.641896e+01 0.000000e+00 0.000000e+00
## ENKUR 0.0000000 9.451048e-01 0.000000e+00 0.000000e+00 0.000000e+00
## ENTPD1 1.6507099 8.935243e-01 1.147919e+01 1.375434e+00 4.898511e+00
## ENY2 26.5002067 2.404341e+01 5.409856e+01 4.986680e+01 3.606147e+01
## EPSTI1 19.0885070 4.731952e+01 7.617608e+01 2.845584e+01 4.398510e+01
## ERICH1 17.3302606 1.175472e+01 1.576855e+01 1.116412e+01 2.458191e+01
## ERP44 9.6108226 1.226388e+01 2.339284e+01 2.007760e+01 2.170484e+01
## ESAM 2.1253536 0.000000e+00 6.045339e-01 7.990087e-01 0.000000e+00
## ETV6 2.5768405 1.494083e+00 1.122636e+01 4.752574e+00 2.447906e+00
## FABP5 23.3418637 1.711111e+01 1.360960e+01 3.002133e+01 2.403654e+01
## FAM118B 4.7082373 5.058657e+00 8.364689e+00 6.934317e+00 4.875352e+00
## FAM133B 24.1431111 2.980681e+01 1.745940e+01 3.143099e+01 2.812259e+01
## FAM89B 70.4331938 8.272632e+01 8.995449e+01 8.620721e+01 2.477784e+01
## FBP1 24.3921414 6.501524e+00 9.387376e+01 1.851556e+01 2.162156e+01
## FCN1 21.4749431 1.528438e+01 7.582849e+02 1.321331e+01 1.931194e+01
## FDFT1 24.5460872 2.870289e+01 2.717549e+01 2.358084e+01 4.866579e+01
## FERMT3 14.1662877 2.530938e+01 2.052459e+01 4.182378e+01 1.436709e+01
## FGL2 5.9780627 5.450367e+00 9.101656e+01 5.321555e+00 6.332911e+00
## FHL1 24.1322086 9.583696e+00 2.021357e+00 7.309011e+00 6.485726e+00
## FIBP 43.4648249 5.111205e+01 1.000041e+02 4.951752e+01 4.537918e+01
## FKBP11 42.1341366 5.798362e+01 5.275944e+00 6.535784e+01 1.353121e+01
## FKBP15 1.6687669 1.299870e+00 5.603379e+00 2.761562e+00 2.115771e+00
## FLNA 15.7702936 2.392243e+01 3.764489e+01 4.350910e+01 1.195616e+01
## FLT3 0.3907995 0.000000e+00 1.403693e+00 0.000000e+00 0.000000e+00
## FOLR3 1.4012956 4.349181e+00 3.568367e+02 2.883298e+00 0.000000e+00
## FUCA2 8.5199521 1.225102e+01 2.839529e+01 1.092367e+01 6.272579e+00
## FUOM 41.7378443 2.970639e+01 1.516408e+02 4.990452e+01 4.286943e+01
## FUT7 7.0822541 9.309786e+01 1.470784e+01 3.843848e+01 2.516254e+01
## FYN 26.1315598 3.999966e+01 1.628328e+01 6.624500e+01 4.581360e+00
## G6PD 11.4394870 9.435020e+00 1.941930e+01 2.887792e+01 1.698879e+01
## GAPDH 999.2657836 1.589479e+03 2.803099e+03 1.687341e+03 7.927222e+02
## GATA3 49.4417950 6.489405e+01 1.699226e+00 4.838802e+01 5.789090e+00
## GDI2 83.4207947 1.170182e+02 1.085700e+02 8.889615e+01 1.666165e+02
## GFI1B 0.0000000 0.000000e+00 4.563082e-01 0.000000e+00 0.000000e+00
## GHITM 70.8059003 8.536869e+01 6.822482e+01 1.095500e+02 1.153520e+02
## GIMAP4 539.9993972 7.078802e+02 2.921802e+02 5.153986e+02 5.722751e+01
## GIMAP5 183.7148424 1.356806e+02 1.824652e+01 1.740599e+02 1.141341e+01
## GIMAP7 1452.8464013 1.715630e+03 7.811940e+02 1.433377e+03 9.650664e+01
## GLA 2.3721609 2.074585e+00 8.800145e+00 2.578171e+00 5.094550e+00
## GLIPR1 25.4669865 3.727600e+01 6.866786e+01 3.693917e+01 4.460408e+01
## GLIPR2 48.2126370 9.531648e+01 1.909620e+02 9.126205e+01 2.140368e+01
## GNB2 60.6276674 1.742095e+02 1.792269e+02 1.083871e+02 1.000930e+02
## GNG11 0.0000000 1.633570e+00 6.731785e+00 7.357996e+00 1.594496e+00
## GNPTAB 2.6814721 4.701416e+00 3.934854e+00 9.801694e+00 8.442566e+00
## GNS 4.3830917 2.483919e+00 2.050798e+01 4.801780e+00 6.223660e+00
## GOLGA7 40.9744568 3.609865e+01 1.271134e+01 2.473028e+01 1.411387e+01
## GPR107 0.7472097 6.834339e-01 2.841783e+00 1.940493e+00 6.432148e-01
## GRINA 16.2347849 2.608630e+01 1.134324e+02 1.565871e+01 2.490869e+01
## GSN 1.4885273 7.243897e-01 1.088024e+01 2.529328e+00 1.151903e+00
## GSTK1 251.9618861 4.133515e+02 2.272010e+02 2.614909e+02 1.326777e+02
## GSTO1 166.1637454 2.418655e+02 4.799424e+02 1.651593e+02 1.083164e+02
## GSTP1 205.0670398 2.441430e+02 2.316236e+03 3.519802e+02 3.227349e+02
## GTF3A 313.8585291 4.325576e+02 2.121678e+02 4.006645e+02 4.037320e+02
## GUSB 22.8386905 3.828583e+01 3.248077e+01 2.871205e+01 1.476155e+01
## HBS1L 6.3902339 3.015496e+00 1.468575e+00 2.553374e+00 1.723068e+00
## HDDC2 37.4418428 3.705288e+01 2.111918e+01 2.360694e+01 2.496604e+01
## HEBP1 9.7114126 1.358354e+01 5.275341e+01 6.658364e+00 8.301655e+00
## HEBP2 5.6680074 7.227521e+00 1.056850e+01 3.141160e+00 7.876186e+00
## HEMGN 8.2300535 2.236110e+00 6.700298e-01 0.000000e+00 0.000000e+00
## HHEX 3.7196144 7.718116e+00 5.831156e+01 7.646391e+00 1.537947e+02
## HNRNPA2B1 115.0249733 1.221514e+02 5.473933e+01 1.349246e+02 1.484313e+02
## HNRNPF 158.6902417 2.840171e+02 9.723659e+01 2.607799e+02 1.661312e+02
## HOTAIRM1 14.8088503 2.013456e+01 9.447886e+01 4.727982e+01 1.166623e+01
## HSP90B1 25.1800522 3.118818e+01 1.737837e+01 4.245350e+01 2.337989e+01
## HSPA5 35.9874134 5.504891e+01 1.273039e+01 7.935598e+01 3.245246e+01
## HSPA8 439.0906959 4.818406e+02 1.433949e+02 4.427268e+02 2.420459e+02
## HVCN1 10.9230543 1.247862e+01 3.435357e+01 6.542968e+01 3.224712e+02
## IDH3G 69.4793465 8.441461e+01 1.587556e+02 1.074832e+02 9.629472e+01
## IDS 39.9812303 5.431279e+01 2.723310e+01 4.503091e+01 3.757786e+01
## IFIT1 35.5121100 3.048467e+01 3.362700e+01 8.356809e+00 1.051914e+01
## IFIT2 6.6072648 1.009147e+01 3.580469e+01 1.052249e+01 6.695138e+01
## IFIT3 18.4087825 3.671996e+01 3.353339e+01 1.332281e+01 3.425551e+01
## IFITM1 740.9355971 9.091839e+02 1.087932e+02 8.850958e+02 2.497907e+02
## IFITM2 1002.8193159 1.221981e+03 1.049586e+03 1.392102e+03 8.871903e+02
## IFITM3 25.3327019 2.305844e+01 6.115072e+02 3.797065e+01 1.988687e+01
## IFNG 7.0182649 1.096792e+01 1.772520e+00 9.461817e+01 1.063119e+01
## IL13RA1 1.6274892 0.000000e+00 1.380237e+01 0.000000e+00 5.557526e+00
## IL18 0.0000000 7.031257e-01 2.303576e+01 0.000000e+00 0.000000e+00
## IL2RG 311.4454144 3.995529e+02 5.011784e+01 4.685806e+02 3.006195e+02
## IL3RA 0.0000000 0.000000e+00 4.014971e+00 2.022745e+00 8.608878e+00
## ILK 28.0263776 2.701553e+01 4.654902e+01 4.355569e+01 3.064793e+01
## IMPDH1 7.9144439 1.662107e+01 5.345394e+01 2.945085e+01 1.096351e+01
## INSIG1 21.6844931 1.598401e+01 7.495601e+00 1.434462e+01 2.136585e+01
## IRF5 0.8576745 2.460070e+00 9.784463e+00 2.425057e+00 6.501345e+00
## IRF7 43.9287664 6.731341e+01 1.019127e+02 1.094008e+02 9.133319e+01
## ITGB7 39.1957020 8.950525e+01 8.169964e+00 6.732662e+01 3.029645e+01
## ITM2A 185.7676129 1.814812e+02 7.172243e+00 1.150423e+02 1.704773e+01
## ITM2B 230.9550095 2.744704e+02 1.722451e+02 2.288131e+02 1.845689e+02
## KCNQ1 4.3215747 5.856782e+00 6.816828e+00 1.009530e+00 4.457610e+00
## KCTD12 3.0039959 1.862408e+00 9.769377e+00 1.900583e+00 4.621468e+00
## KLF10 7.5937183 8.532820e+00 1.482784e+01 9.700546e+00 6.509115e+00
## KLF4 6.1042311 1.149520e+01 1.347259e+02 7.970812e+00 1.318050e+01
## KLF6 208.3857664 3.946525e+02 3.104919e+02 3.762818e+02 2.332419e+02
## KLHL42 3.9706544 2.625155e+00 2.787500e+00 4.306908e+00 1.930980e+00
## KLRB1 33.6391239 1.121879e+02 7.089834e+00 1.978655e+02 0.000000e+00
## KLRC1 0.0000000 9.018126e+00 5.043767e-01 3.558414e+01 1.131622e+00
## KLRD1 0.6287306 8.882844e-01 7.714806e-01 1.651070e+01 0.000000e+00
## KLRF1 9.7946107 5.954787e+00 1.144926e+00 2.482855e+01 3.582213e+00
## KLRG1 66.2753041 5.666985e+01 6.980903e+00 3.848327e+02 1.234245e+01
## LAG3 2.2156374 1.382584e+01 6.487856e-01 9.880602e+01 0.000000e+00
## LAMTOR1 163.4623072 1.500527e+02 3.024311e+02 1.755272e+02 2.050010e+02
## LAMTOR4 396.1437425 3.930000e+02 7.933482e+02 4.400311e+02 4.657076e+02
## LAT2 5.8097033 4.960207e+00 3.359033e+01 6.599621e+00 1.463875e+02
## LCN2 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## LCP1 92.0631606 1.277365e+02 1.094074e+02 1.617978e+02 8.571927e+01
## LDHA 115.4279121 1.862180e+02 1.230763e+02 1.869117e+02 9.591065e+01
## LDHB 1337.2921806 1.362879e+03 1.930559e+02 6.790674e+02 2.759276e+02
## LEPROTL1 129.4591193 1.278794e+02 2.540820e+01 6.702011e+01 2.341049e+01
## LFNG 6.7942144 4.210043e+00 2.952658e+01 5.200948e+00 4.571736e+00
## LIMK1 0.0000000 9.933688e-01 3.204206e+00 3.304719e+00 2.388635e+00
## LMO2 0.8215086 1.585306e+00 2.338451e+01 2.851977e+00 2.199172e+00
## LRP1 0.4448904 7.473859e-01 5.315199e+00 5.665666e-01 3.222821e-01
## LSP1 138.1116608 1.430230e+02 1.667477e+02 2.018667e+02 1.631668e+02
## LTA4H 23.0394235 1.494178e+01 4.243855e+01 1.662761e+01 5.723024e+01
## LTV1 56.3858576 7.389741e+01 1.295972e+01 3.073593e+01 4.165338e+01
## LY6E 472.5162456 6.952436e+02 9.706697e+02 5.585959e+02 5.669875e+02
## LYN 5.3763297 1.087688e+01 4.993741e+01 1.289059e+01 7.234146e+01
## LYPD2 2.7355075 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## LYZ 506.9093205 5.087432e+02 2.357145e+04 4.792729e+02 5.414975e+02
## M6PR 82.5454809 1.001183e+02 6.311996e+01 8.111838e+01 5.589051e+01
## MAD1L1 17.5200485 1.596886e+01 1.120147e+01 2.697917e+01 1.828423e+01
## MAF1 112.1314208 1.366784e+02 7.168693e+01 1.252297e+02 2.727575e+02
## MAP3K11 4.1835480 2.972117e+00 1.262781e+01 5.253660e+00 6.064675e+00
## MAP3K8 3.1376466 4.408338e+00 2.215628e+01 1.786874e+01 1.056411e+02
## MEST 1.0971957 1.662360e+00 0.000000e+00 0.000000e+00 1.825573e+00
## MFSD3 9.0829451 5.283096e+00 6.020048e+00 4.911754e+00 1.119561e+01
## MGST1 0.0000000 1.031817e+00 2.143836e+01 0.000000e+00 0.000000e+00
## MICAL1 7.6566388 4.596994e+00 1.080504e+01 4.627357e+00 1.228573e+01
## MLEC 8.7488302 1.498898e+01 1.143332e+01 1.228961e+01 1.442306e+01
## MOB3B 0.0000000 2.988092e-01 1.060387e+00 0.000000e+00 3.039416e+00
## MPC1 116.5418646 1.328662e+02 1.352659e+02 1.228642e+02 6.430110e+01
## MPEG1 4.8513247 4.587725e+00 9.135894e+01 2.850586e+00 2.476464e+01
## MPP1 7.0738201 4.899642e+00 2.614429e+01 2.827393e+00 2.765002e+00
## MRPL18 200.3234141 1.583053e+02 1.373133e+02 2.005629e+02 1.791513e+02
## MRPL21 76.8435327 5.649020e+01 7.308405e+01 1.307284e+02 6.963172e+01
## MRPL23 116.4858942 1.397670e+02 1.944190e+02 1.027454e+02 1.067339e+02
## MS4A1 11.3075188 1.216381e+01 9.975800e+00 1.722353e+01 7.227630e+02
## MS4A4A 0.5730596 1.538275e+00 4.432575e+00 1.521032e+00 4.741992e-01
## MS4A6A 4.3493602 5.419208e+00 3.151759e+02 5.905733e+00 4.429356e+00
## MS4A7 5.0701518 6.353786e+00 7.738004e+01 3.824049e+00 2.291274e+01
## MSANTD3 2.7489481 2.472985e+00 8.895645e-01 2.631330e+00 2.449278e+00
## MSN 26.0708149 2.948727e+01 3.795263e+01 3.592034e+01 2.533953e+01
## MSRA 20.0969807 1.399836e+01 2.170878e+01 7.263409e+00 1.662288e+01
## MSRB2 24.4395637 2.601598e+01 8.991427e+01 1.005491e+01 7.214048e+00
## MTIF3 64.5766568 1.644184e+02 8.628348e+01 8.467447e+01 7.581589e+01
## MTPN 34.1762441 3.103846e+01 4.575568e+01 2.958425e+01 7.626214e+01
## MTSS1 8.7067675 8.286170e+00 1.193355e+01 1.150895e+01 1.948340e+01
## MTURN 0.3223213 4.641905e-01 1.803029e+00 0.000000e+00 1.690748e+00
## MYC 70.1062891 5.125286e+01 6.518644e+00 3.014108e+01 6.343681e+01
## MYL6 440.6035423 6.759596e+02 9.162297e+02 8.042208e+02 4.691024e+02
## MYO1G 22.7363604 3.614810e+01 3.467725e+01 5.270633e+01 2.662400e+01
## MYOF 0.0000000 2.602861e-01 2.329614e+00 0.000000e+00 0.000000e+00
## NAMPT 3.3208667 2.087420e+00 6.740050e+00 2.909524e+00 2.177261e+00
## NANS 20.7012863 1.379673e+01 4.933748e+01 2.637433e+01 1.391970e+01
## NAP1L1 86.7996371 9.205015e+01 3.891414e+01 4.970610e+01 9.474277e+01
## NCF1 31.9650405 2.820048e+01 1.779759e+02 1.595228e+01 2.588265e+02
## NCKAP1L 8.0742076 9.268023e+00 2.080226e+01 1.077554e+01 1.081865e+01
## NCOA4 37.2312382 5.010099e+01 4.418374e+01 3.806414e+01 3.245662e+01
## NDUFB11 156.8336905 2.196548e+02 2.247817e+02 1.998769e+02 1.897316e+02
## NDUFB2 112.2726723 1.107423e+02 1.178666e+02 1.151611e+02 7.939420e+01
## NEAT1 50.0687973 5.783392e+01 1.395650e+02 5.954471e+01 3.357809e+01
## NECAP1 7.3240739 6.273952e+00 5.280256e+00 8.970869e+00 1.109284e+01
## NEK3 0.4429219 3.440522e+00 2.578851e+00 9.774986e-01 3.148670e+00
## NELL2 38.9451281 2.197699e+01 2.017010e+00 1.086654e+01 0.000000e+00
## NEURL1 3.5433525 2.201127e+00 2.154833e+00 0.000000e+00 1.942823e-01
## NFE2 0.7009498 0.000000e+00 5.361605e+01 1.334406e+00 0.000000e+00
## NINJ1 48.3763932 5.244222e+01 1.382699e+02 4.042200e+01 1.134340e+01
## NIPSNAP3A 17.6975396 1.476478e+01 4.530988e+01 1.986853e+01 2.516309e+01
## NOC4L 32.6339180 6.315368e+01 1.992733e+01 2.111282e+01 2.339636e+01
## NR4A1 11.0747516 1.432748e+01 3.418253e+01 7.860794e+00 1.615963e+01
## NRG1 0.0761251 2.770593e-01 7.812032e+00 2.541474e-01 2.764609e-01
## NRGN 3.6855367 1.853685e+00 9.154177e+01 0.000000e+00 1.242925e+01
## NSMF 1.7508784 1.605812e+00 7.327664e+00 2.893854e+00 1.232718e+00
## NT5C3A 73.4165414 3.671773e+01 4.497160e+01 2.504736e+01 6.942702e+01
## NUCB2 36.8451148 1.704095e+01 4.303092e+00 3.115552e+01 5.618161e+00
## NUP214 1.7130999 1.652193e+00 3.164566e+01 1.850200e+00 2.279602e+00
## OAS1 17.3243438 1.655310e+01 2.791280e+01 1.075302e+01 2.478560e+01
## OAS3 10.1775265 4.573499e+00 6.359129e+00 3.052689e+00 2.854542e+00
## OPTN 32.7918251 8.737962e+01 5.494700e+00 7.129960e+01 1.329726e+01
## ORAI2 4.8868314 4.985405e+00 6.175740e+00 6.638461e+00 3.642464e+01
## OS9 13.4491079 1.806103e+01 2.008683e+01 2.236615e+01 1.690418e+01
## OSTF1 281.7291060 3.005826e+02 3.529393e+02 3.766427e+02 2.723388e+02
## PAK1 3.1179773 2.823439e+00 2.232613e+01 5.013066e-01 4.599881e+00
## PCGF5 11.7613363 1.519606e+01 1.925205e+01 1.866837e+01 1.538899e+01
## PDLIM1 2.6866632 8.485779e+00 4.988586e+00 1.235890e+01 2.665518e+02
## PEBP1 568.1133041 5.992169e+02 1.272699e+02 5.207719e+02 4.652780e+02
## PGAM1 79.2908204 1.051852e+02 1.502717e+02 1.426033e+02 9.238233e+01
## PGK1 139.9819282 1.420727e+02 1.308124e+02 1.538949e+02 1.342399e+02
## PGRMC1 25.9910896 1.551668e+01 2.076270e+01 2.526973e+01 1.264052e+01
## PHF19 3.8379332 6.335127e+00 9.280878e+00 5.214480e+00 1.820154e+00
## PHLDA2 3.8952356 9.311545e+00 4.569577e+01 3.454516e+00 1.165252e+01
## PHPT1 85.8635699 1.109413e+02 1.990608e+02 1.888208e+02 8.100949e+01
## PIK3AP1 1.5993684 4.284299e+00 1.455902e+01 8.551701e+00 1.494591e+01
## PILRA 6.1945265 7.887298e+00 1.888263e+02 1.387894e+01 1.348569e+01
## PKIB 0.0000000 0.000000e+00 7.043887e-01 0.000000e+00 1.334167e+00
## PLBD1 7.7010540 9.547684e-01 2.362181e+02 9.666134e+00 9.165090e+00
## PLEKHF2 19.3012198 1.932079e+01 1.457189e+01 2.131291e+01 1.637538e+02
## PLIN2 26.3322962 3.904443e+01 2.088406e+01 3.376947e+01 9.887383e+00
## PLP2 406.4594282 8.124533e+02 4.368757e+02 3.060435e+02 4.062461e+02
## PLXNC1 0.4188288 8.323931e-01 2.380189e+00 7.670316e-01 1.902198e+00
## PNISR 69.8518259 5.640121e+01 1.691986e+01 6.978584e+01 1.049362e+02
## PNOC 5.1755211 1.861583e+00 5.890602e+00 2.033757e+00 2.016994e+02
## PNPLA4 9.1826584 6.059553e+00 5.319751e+00 4.475523e+00 8.622680e+00
## POLD2 11.4138409 1.388499e+01 5.755462e+00 8.296064e+00 1.582355e+01
## POLD4 135.9245101 1.241900e+02 1.769453e+02 1.341463e+02 3.686721e+02
## POLR2G 179.6096878 1.546904e+02 2.009394e+02 1.850495e+02 1.896102e+02
## POLR2L 406.2056733 4.360740e+02 6.772672e+02 4.734286e+02 2.987921e+02
## POM121 3.9182760 4.222550e+00 3.368705e+00 6.002569e+00 1.021911e+01
## POMP 29.2899696 3.006736e+01 3.437882e+01 4.735452e+01 4.108735e+01
## PON2 2.1953844 1.012619e+00 2.202854e+00 6.279797e+00 0.000000e+00
## POU2AF1 1.6799853 1.173342e+00 0.000000e+00 0.000000e+00 4.103088e+01
## PPA1 132.7009197 1.396072e+02 4.762847e+01 8.887367e+01 7.954053e+01
## PPIA 245.8616258 2.666392e+02 1.812252e+02 3.331508e+02 3.047821e+02
## PPP1CA 153.8421746 2.165507e+02 1.990939e+02 2.775097e+02 2.162830e+02
## PPP1R12A 13.7177491 2.688251e+01 7.540123e+00 2.262174e+01 1.878594e+01
## PPP1R14B 11.9071662 1.491222e+01 7.770378e+00 1.407114e+01 2.481180e+01
## PQBP1 55.8285794 5.437652e+01 1.246343e+02 6.122523e+01 4.478898e+01
## PRDX3 28.2627714 7.599109e+01 6.061463e+01 7.286318e+01 4.673490e+01
## PRDX5 433.0913810 4.917144e+02 8.306309e+02 6.798904e+02 5.637604e+02
## PRF1 25.9014073 4.062252e+01 1.357803e+01 4.979479e+02 1.777129e+01
## PRKAR2B 0.3461950 1.584357e-01 6.880405e-02 0.000000e+00 0.000000e+00
## PRKCQ-AS1 108.8536101 6.343923e+01 1.955360e+00 4.347157e+01 2.220180e+00
## PRR13 242.0241147 2.813499e+02 2.857726e+02 3.376094e+02 3.281054e+02
## PRSS23 1.0914015 1.509841e-01 2.834963e-01 1.079206e+01 8.113425e-01
## PSAP 159.8138498 2.458528e+02 1.224941e+03 1.781187e+02 2.169399e+02
## PSIP1 61.2366522 5.461392e+01 1.233214e+01 3.992310e+01 3.705843e+01
## PSMB1 319.5874450 3.266792e+02 3.104316e+02 3.784737e+02 3.372950e+02
## PSMC3 77.4482965 1.025144e+02 6.681770e+01 1.178137e+02 8.757254e+01
## PSMD9 23.2901452 2.687097e+01 2.995534e+01 3.017317e+01 1.963836e+01
## PSMG3 47.9946194 3.648945e+01 3.996476e+01 4.533526e+01 2.991616e+01
## PTGS1 0.3109435 0.000000e+00 3.303573e+00 1.179526e+00 4.184570e+00
## PTK2 1.0065873 2.755428e-01 3.313837e-01 4.875261e-01 1.864787e+00
## PTMS 4.6380829 7.454265e+00 2.557561e+01 6.168254e+01 0.000000e+00
## PTP4A3 15.0041853 1.101331e+01 7.213090e+00 5.130461e+00 3.129805e+00
## PTPN6 52.0541337 5.694535e+01 8.553306e+01 7.735867e+01 1.582762e+02
## PTPRCAP 2364.8041349 2.724271e+03 1.375136e+02 4.672600e+03 4.102082e+03
## PTPRE 4.2578102 4.928085e+00 2.499679e+01 5.996601e+00 9.440009e-01
## QRSL1 10.3971881 7.155869e+00 2.927938e+00 6.725257e+00 9.934479e+01
## RAB18 9.7847170 1.233393e+01 6.629695e+00 1.622138e+01 8.680034e+00
## RAB32 1.9354637 1.520223e+01 3.826323e+02 1.555250e+01 1.252593e+01
## RAC1 46.6932130 7.429710e+01 2.608846e+02 6.317421e+01 5.099333e+01
## RAN 199.2281220 2.371826e+02 1.115944e+02 2.003803e+02 1.727808e+02
## RAP1B 36.2163293 2.832018e+01 1.613186e+01 5.054594e+01 2.154722e+01
## RASSF4 0.8460876 9.393432e-01 1.945669e+01 1.079085e+00 2.862080e+00
## RELT 3.3830794 2.264645e+00 1.425494e+01 2.894785e+00 1.348999e+01
## REPIN1 16.7195102 1.139232e+01 1.179461e+01 9.862102e+00 2.231383e+01
## RGCC 653.1556302 6.532765e+02 2.196568e+02 3.057335e+02 7.602004e+01
## RGS10 950.9844813 6.458235e+02 5.183434e+02 4.165761e+02 1.069412e+02
## RGS3 0.8124385 2.565413e+00 2.042062e+00 5.095018e+00 3.854997e-01
## RHOBTB1 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## RHOG 197.5066769 2.979389e+02 6.376110e+02 3.661389e+02 1.828438e+02
## RILPL2 89.2068270 1.713062e+02 2.838181e+02 1.335156e+02 1.088399e+02
## RIN1 1.3302805 1.544577e+00 9.815676e+00 1.985084e+00 0.000000e+00
## RMDN1 10.1387491 4.889034e+00 6.940371e+00 7.613493e+00 8.123471e+00
## RNASEH2B 6.2159396 9.149019e+00 7.836578e+00 9.197655e+00 1.701955e+01
## RNASET2 39.3448961 5.866143e+01 4.430284e+01 2.707782e+01 6.766007e+01
## RNH1 49.4160985 5.066686e+01 8.626053e+01 4.265919e+01 4.443562e+01
## RPS6KA4 3.9676565 8.090593e+00 2.211550e+01 1.106440e+01 5.199347e+00
## RPS6KB2 19.0204498 1.799884e+01 1.996664e+01 2.587855e+01 1.808371e+01
## RSU1 32.3199032 4.342401e+01 1.741340e+01 4.275250e+01 3.538568e+01
## RTN3 17.1664695 2.330110e+01 4.392984e+01 2.528161e+01 7.708659e+00
## RWDD1 74.6077555 1.002663e+02 3.671486e+01 6.347208e+01 4.255369e+01
## RXRA 0.8765480 1.722497e+00 1.495218e+01 1.403448e+00 5.067474e-01
## SAMD3 15.3407002 2.317713e+01 1.852569e+00 8.541446e+01 4.981566e+00
## SASH3 26.8758668 4.287784e+01 5.603442e+01 7.466500e+01 6.508307e+01
## SAT1 275.8899589 4.218489e+02 2.495635e+03 4.582140e+02 5.186322e+02
## SBDS 140.7743870 1.498217e+02 3.696994e+01 9.857635e+01 1.400303e+02
## SCGB1C1 0.0000000 0.000000e+00 4.808457e+00 0.000000e+00 0.000000e+00
## SDCBP 48.7557729 7.144303e+01 1.079199e+02 5.343027e+01 5.436237e+01
## SDHAF2 78.6670510 7.550493e+01 4.313704e+01 1.489184e+02 4.433242e+01
## SELPLG 67.9402226 9.580908e+01 6.960943e+01 9.562496e+01 3.127015e+01
## SENCR 1.1881180 3.908495e+00 1.774888e+01 8.579568e+00 0.000000e+00
## SERPINE1 0.8179727 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## SETDB2 8.9587698 5.336848e+00 7.075669e+00 2.077620e+00 1.424730e+01
## SF3B5 743.2914093 1.295286e+03 1.494729e+03 9.962390e+02 7.332760e+02
## SFT2D1 40.3762799 5.519322e+01 5.319814e+01 7.792497e+01 6.447683e+01
## SH2D1A 24.8891033 3.979567e+01 3.250230e+00 1.360431e+02 3.327270e+00
## SH2D3C 8.7098406 6.683629e+00 8.535968e+00 7.931307e+00 8.591418e+00
## SH3BGRL 249.2838720 3.405800e+02 3.869138e+02 2.690010e+02 4.308659e+02
## SH3BGRL2 1.2613223 4.742848e-01 4.367880e-01 1.644508e+00 0.000000e+00
## SIAE 1.1427595 4.642884e-01 3.409580e-01 3.707828e-01 8.133308e-01
## SIPA1 21.8982601 2.040093e+01 1.527479e+01 4.392659e+01 4.111346e+01
## SIT1 203.2519105 3.111659e+02 4.253160e+00 3.070989e+02 1.400572e+02
## SKAP2 2.2908671 7.129968e+00 1.819843e+01 8.856420e+00 2.897594e+01
## SLC25A3 101.0648763 1.051835e+02 9.366630e+01 1.095897e+02 1.053046e+02
## SLC25A5 512.6493158 5.166776e+02 9.694146e+02 5.196177e+02 5.518003e+02
## SLC25A6 1715.8684509 1.754559e+03 1.975512e+03 1.447369e+03 1.989905e+03
## SLC2A3 100.1825355 9.903658e+01 2.441031e+01 3.541282e+01 5.975735e+01
## SLC2A6 3.8419385 2.487481e+00 2.042832e+01 2.461443e+00 2.321046e+01
## SLC31A2 8.1086833 5.730471e+00 1.057788e+02 1.007459e+01 3.401127e+00
## SLC35D2 2.3621124 6.217643e+00 3.076529e+00 4.045081e+00 1.178802e+00
## SMCO4 14.8478863 2.554318e+01 4.357146e+02 1.823367e+01 2.327325e+01
## SMNDC1 4.4354397 1.200005e+01 7.459041e+00 9.711759e+00 1.182646e+01
## SMPDL3A 0.0000000 0.000000e+00 8.821280e+00 0.000000e+00 2.323344e+00
## SMS 49.1598427 8.653337e+01 1.270234e+02 1.092886e+02 1.378444e+02
## SNHG7 103.7053361 6.903546e+01 5.172537e+01 5.874893e+01 3.011167e+02
## SNX10 2.1845278 4.716989e+00 2.146385e+01 7.812573e+00 4.517279e+01
## SNX3 350.4426622 2.709854e+02 5.080750e+02 3.088781e+02 8.902320e+02
## SOD2 10.2019446 1.184782e+01 3.262983e+01 7.290688e+00 9.031168e+00
## SPCS2 126.5837986 1.797965e+02 5.381202e+01 2.087673e+02 2.291482e+02
## SPI1 6.4860694 7.423011e+00 2.920135e+02 1.234716e+01 5.577309e+01
## SPOCK2 71.1944250 1.060279e+02 4.705937e+00 7.133352e+01 2.427312e+01
## SPRYD4 0.4785949 9.519597e-01 1.350350e+00 2.951207e-01 5.250767e-01
## SRGN 541.2020478 9.425576e+02 2.620281e+03 1.785360e+03 2.571716e+02
## SRI 70.6607355 9.472477e+01 6.650922e+01 1.020545e+02 7.217543e+01
## ST14 0.9359978 5.029226e-01 3.691359e+00 0.000000e+00 3.990260e+00
## ST3GAL4 1.9037039 1.884001e+00 2.407389e+00 5.910430e+00 7.431317e-01
## STAC3 4.1241736 4.081175e+00 4.258147e+01 0.000000e+00 6.303355e+00
## STAT2 3.9626857 7.232622e+00 1.055183e+01 5.481676e+00 8.264077e+00
## STIP1 12.3430689 2.340509e+01 8.068955e+00 1.791437e+01 2.191496e+01
## STK32C 2.8578134 1.110343e+01 1.298802e+01 8.472247e+00 3.202324e+00
## STK38L 2.6699061 4.189969e+00 5.329589e+00 3.251878e+00 1.291075e+00
## STOM 14.5221939 5.014321e+01 1.140357e+01 7.603672e+01 6.252227e+00
## STRBP 3.5061971 4.458701e+00 1.504445e-01 1.749245e+00 2.988651e+01
## STX11 3.6460925 4.064466e+00 1.029930e+02 2.310752e+01 3.464573e+00
## STX7 2.4870879 3.904281e+00 1.076801e+01 6.075428e+00 3.257900e+01
## SUSD3 247.2338030 3.501730e+02 5.266828e+01 8.020478e+01 7.760449e+01
## SWAP70 2.5361775 1.928480e+00 3.158954e+00 2.775737e+00 6.215232e+01
## SWI5 18.0141052 2.009380e+01 2.115262e+01 1.819561e+01 3.202761e+01
## SYK 3.1863197 4.460509e+00 3.881272e+01 5.664780e+00 3.402418e+01
## SYNE1 1.4144652 1.393862e+00 3.267545e-01 3.419097e+00 1.305255e+00
## SYPL1 76.8397838 6.106975e+01 2.712800e+01 6.032359e+01 1.906349e+02
## SYTL4 0.0000000 2.302045e-01 0.000000e+00 0.000000e+00 0.000000e+00
## TAF10 31.9342784 8.870937e+00 6.206165e+00 7.233879e+00 6.296539e+00
## TALDO1 120.2045286 1.884563e+02 5.796113e+02 1.556625e+02 1.394203e+02
## TBC1D10C 109.2693550 1.072200e+02 2.726238e+01 1.318705e+02 1.083913e+02
## TBL2 2.5092203 4.134664e+00 2.313552e+00 3.541853e+00 1.407406e+00
## TBXAS1 4.5455206 6.768832e+00 5.770177e+01 5.474769e+00 6.544749e+00
## TCEAL8 133.9428892 2.049549e+02 2.275316e+02 1.382129e+02 1.000018e+02
## TCF7L2 0.2749248 2.048223e-01 7.447599e+00 1.046936e+00 3.150507e+00
## TCIRG1 10.3193012 1.153359e+01 3.640383e+01 1.980465e+01 1.969587e+01
## TDRP 0.0000000 0.000000e+00 7.338304e-01 0.000000e+00 0.000000e+00
## TES 24.2195132 2.169242e+01 1.301647e+01 3.740672e+01 2.326968e+01
## TESC 10.7121830 1.206396e+01 8.971089e+01 3.834388e+01 1.714528e+01
## TIMM8B 76.1408489 8.576925e+01 1.675413e+02 1.073069e+02 7.161352e+01
## TIMP1 175.4298259 4.030913e+02 8.805650e+02 2.025749e+02 5.191924e+01
## TJP2 0.3336283 2.982399e-01 0.000000e+00 4.055535e-01 0.000000e+00
## TLN1 6.9675862 9.928958e+00 2.252001e+01 1.792506e+01 1.581132e+01
## TMEM123 257.4576415 2.247636e+02 7.933985e+01 1.510445e+02 1.626224e+02
## TMEM134 28.8312701 2.551795e+01 1.984543e+01 1.451186e+01 3.648750e+01
## TMEM140 13.4124872 1.900577e+01 1.397557e+01 1.402953e+01 1.022102e+01
## TMEM141 78.4053044 8.075979e+01 1.045270e+02 6.893746e+01 6.765236e+01
## TMEM176A 0.4913541 9.029162e-01 3.229631e+01 8.650240e-01 3.012772e+00
## TMEM176B 3.1891596 4.156764e+00 1.901157e+02 1.604933e+00 5.541212e+00
## TMEM179B 70.7218103 8.699757e+01 1.748858e+02 1.422308e+02 1.375557e+02
## TMEM218 5.3161707 6.762962e+00 6.770344e+00 5.222020e+00 6.763777e+00
## TMEM243 98.1477340 1.011005e+02 4.143109e+01 4.954923e+01 1.678598e+02
## TMPO 10.1896138 9.832551e+00 8.788296e+00 7.489683e+00 1.106810e+01
## TNFAIP3 37.4259275 6.841010e+01 1.842193e+01 4.198574e+01 1.844800e+01
## TNFRSF1A 10.0367322 1.728222e+01 5.988899e+01 3.466772e+01 0.000000e+00
## TNNI2 0.0000000 0.000000e+00 7.240172e+01 2.966916e+00 2.250100e+01
## TOLLIP 8.4136412 1.236313e+01 2.096822e+01 1.240356e+01 7.642207e+00
## TOMM7 1690.4129781 1.491873e+03 8.454105e+02 1.215584e+03 1.251290e+03
## TPD52 7.7566770 1.230920e+01 2.160283e+00 1.416995e+01 5.846293e+01
## TPI1 219.1643970 3.281820e+02 5.122144e+02 3.044981e+02 2.175731e+02
## TPP1 12.5495247 1.421845e+01 1.782839e+01 1.675540e+01 2.225563e+01
## TSC22D1 1.6993328 2.380292e+00 1.381883e+00 4.535693e+00 3.879613e+00
## TSC22D3 515.9608299 6.139674e+02 2.127797e+02 5.178735e+02 6.333391e+02
## TSPAN13 1.4000137 5.169899e+00 1.931142e+00 5.789894e+00 3.260817e+02
## TSPAN14 8.5338330 8.427076e+00 6.198444e+00 6.616419e+00 2.388211e+00
## TSPAN18 2.7554314 3.597615e+00 0.000000e+00 1.399500e+00 0.000000e+00
## TSPAN33 0.4881877 0.000000e+00 2.525804e+00 2.920643e+00 1.259642e+01
## TTYH3 0.9256392 9.426363e-01 1.523602e+01 1.129604e+00 6.382517e-01
## TUBA1A 249.9818498 2.275443e+02 2.194157e+02 1.743633e+02 2.249792e+02
## TUBA1B 196.4947051 2.101549e+02 1.394181e+02 1.901583e+02 2.236512e+02
## TUBA1C 13.7246917 1.385148e+01 1.585030e+01 1.898409e+01 8.103950e+00
## TXN 221.2678332 4.071599e+02 2.503097e+02 3.631209e+02 1.177329e+02
## UBC 660.0458287 9.671638e+02 5.766416e+02 1.045330e+03 7.908529e+02
## UBE2A 14.6354710 2.378223e+01 2.020124e+01 3.106464e+01 3.101626e+01
## UBE2D1 22.9636371 2.643681e+01 1.027708e+02 3.240234e+01 2.518718e+01
## UBE2J1 13.8046912 2.872585e+01 4.081141e+01 2.729802e+01 6.711109e+01
## UBE2L6 146.7145248 2.425012e+02 3.826846e+02 2.292549e+02 3.482872e+02
## UBE2R2 9.7708153 1.126129e+01 3.495807e+01 9.495179e+00 1.366347e+01
## UBL4A 28.0179849 3.685986e+01 1.524769e+01 3.434118e+01 2.058737e+01
## UCHL3 46.9455362 4.486076e+01 4.220713e+01 2.640182e+01 3.531759e+01
## UCP2 339.7965624 3.710049e+02 2.921816e+02 4.391600e+02 6.616822e+02
## UNC93B1 6.6834841 1.184030e+01 4.853115e+01 1.247600e+01 4.602511e+01
## UPF3B 34.7544386 4.628954e+01 1.104847e+01 5.074422e+01 7.072518e+01
## UPP1 34.5426964 5.034011e+01 4.645395e+01 5.202426e+01 3.791784e+00
## UTRN 5.2552971 5.898649e+00 5.684457e+00 7.926323e+00 7.393552e+00
## VCL 2.3526523 3.764687e+00 4.094915e+00 5.749409e+00 3.208320e+00
## VIM 582.0139324 9.902642e+02 1.132686e+03 5.222619e+02 3.255358e+02
## VMA21 13.1039733 1.181613e+01 3.612574e+01 1.065696e+01 1.435667e+01
## VPS29 77.4705349 1.054595e+02 6.133286e+01 1.046422e+02 4.753596e+01
## WAS 72.9202899 7.583838e+01 8.965154e+01 7.850204e+01 8.070509e+01
## WNT11 0.0000000 0.000000e+00 0.000000e+00 1.845808e+00 0.000000e+00
## XPNPEP1 6.0898682 6.029742e+00 6.143840e+00 8.777742e+00 8.326304e+00
## YBX3 5.8883832 3.325888e+00 3.331932e+01 6.925980e+00 4.642023e+01
## YIF1A 42.2436379 3.666882e+01 4.986461e+01 5.317125e+01 4.253312e+01
## YTHDF3 6.0451366 5.306795e+00 2.948025e+00 7.390970e+00 6.718356e+00
## YWHAZ 69.7429419 8.277449e+01 4.033046e+01 6.843647e+01 6.209906e+01
## ZBTB16 0.5087594 6.799454e-01 8.417090e-01 4.304802e+00 1.802301e+00
## ZCCHC7 17.0175603 2.081485e+01 6.184247e+00 1.451254e+01 7.275673e+01
## ZFAND5 30.9263232 2.403025e+01 4.238441e+01 2.200458e+01 1.568008e+01
## ZNF385A 3.1030723 4.253310e+00 4.298040e+01 3.441388e+00 7.326392e+00
## ZNF398 3.8523129 1.274687e+00 3.082432e+00 0.000000e+00 5.499281e-01
## ZNF467 4.5619369 2.017208e+00 6.018527e+01 1.073703e+00 4.284111e+00
## ZNF503 0.0000000 0.000000e+00 1.202081e+01 0.000000e+00 0.000000e+00
## ZNF703 2.7004592 1.408339e+00 2.070406e+01 2.495764e+00 2.752226e+00
## ZNF706 54.5455174 5.297099e+01 6.827155e+01 5.101759e+01 6.943333e+01
## ZNF789 1.5437341 2.455570e+00 1.284942e+00 8.404164e-01 3.904744e+00
## ZNHIT1 51.9047848 7.318897e+01 1.076057e+02 7.336583e+01 4.262980e+01
## ZYX 36.7088624 7.348042e+01 1.001946e+02 4.440234e+01 1.288182e+01
## AATK 0.0000000 0.000000e+00 2.934676e-01 0.000000e+00 0.000000e+00
## ABCC3 0.0000000 0.000000e+00 1.091958e+00 0.000000e+00 0.000000e+00
## ABHD17A 7.2025458 1.259998e+01 5.108799e+00 3.815139e+01 6.253399e+00
## ABI3 30.2518338 4.001419e+01 1.246199e+02 1.526616e+02 3.053215e+01
## ACAA2 16.9686976 1.678823e+01 1.912300e+01 3.019767e+01 1.110301e+01
## ACAP1 116.8433332 9.252595e+01 9.065436e+00 1.175249e+02 6.435325e+01
## ACO2 7.0745291 8.610452e+00 7.110491e+00 9.403274e+00 1.542890e+01
## ACTG1 767.4978636 1.173814e+03 8.083578e+02 1.267887e+03 6.695208e+02
## ACTN1 15.0053498 5.708673e+00 5.075507e+00 2.521261e+00 8.725086e-01
## ADA 7.7466916 1.313500e+01 2.343221e+00 1.248416e+01 9.743521e+00
## ADAP2 2.4121190 2.090319e+00 2.519773e+01 1.476251e+00 8.706752e+00
## ADCY7 6.1049162 4.789188e+00 4.859209e+00 7.471637e+00 3.421418e+00
## AHSA1 50.5011371 6.647553e+01 4.564994e+01 7.205472e+01 4.767219e+01
## ALDOA 383.5907234 4.353196e+02 6.401332e+02 3.744035e+02 2.843518e+02
## ALOX12 0.7957774 1.974158e+00 6.121651e-01 2.640683e+00 0.000000e+00
## ANAPC11 68.0581683 9.717701e+01 1.040563e+02 6.638889e+01 5.564884e+01
## ANKRD12 46.7639140 4.793674e+01 1.130093e+01 4.320689e+01 4.414927e+01
## ANKRD9 0.0000000 3.584942e-01 8.992857e-01 1.269464e+00 0.000000e+00
## ANXA2 16.1540107 7.074669e+01 1.590535e+02 7.788175e+01 2.838133e+01
## AP2A1 4.5048367 3.383666e+00 1.296587e+01 6.191435e+00 6.059995e+00
## AP2S1 169.1395118 1.940732e+02 9.167784e+02 2.354059e+02 1.699220e+02
## APEX1 190.8874091 2.140354e+02 1.356503e+02 1.559994e+02 2.503088e+02
## APH1B 4.9605346 6.584551e+00 1.253037e+01 1.192319e+01 9.077637e+00
## APMAP 62.6292531 4.771295e+01 2.936066e+01 2.039862e+02 3.634330e+01
## APOBEC3A 8.1250149 1.904745e+01 4.480944e+02 7.026083e+00 7.846473e+00
## APOBEC3B 0.9767370 2.591128e+00 1.375279e+02 4.856687e+00 6.459954e+00
## APOBEC3G 30.9188375 4.264434e+01 1.928848e+01 2.124225e+02 5.805492e+01
## APOBR 4.0920282 1.037911e+01 2.025081e+01 7.376713e+00 7.478249e-01
## ARG2 0.8818959 8.591691e-01 6.649187e-01 3.237657e+00 1.227012e+00
## ARHGAP27 2.3127379 3.268548e+00 1.157022e+01 4.175508e+00 5.287851e+00
## ARHGDIA 112.0853617 2.145516e+02 1.292920e+02 1.597540e+02 1.764222e+02
## ARHGEF40 0.6069639 1.574883e+00 2.484697e+01 5.475033e-01 1.057585e+00
## ARL6IP1 71.8555790 8.137490e+01 3.051786e+01 1.013873e+02 7.641887e+01
## ARRB2 44.9884434 6.493502e+01 1.818881e+02 7.873223e+01 2.688188e+01
## ASGR1 0.3643140 8.164232e-01 1.448178e+02 0.000000e+00 1.244743e+00
## ATP2A3 13.2846734 1.386788e+01 3.018672e+00 1.426455e+01 2.362148e+01
## ATP6V0D1 8.9393255 1.507260e+01 5.291020e+01 2.577825e+01 1.903713e+01
## ATP9A 0.2358829 0.000000e+00 0.000000e+00 0.000000e+00 6.851643e-01
## BAX 91.9190991 1.411636e+02 1.702674e+02 2.020217e+02 1.640941e+02
## BCL2A1 50.0871484 3.134224e+01 3.925146e+01 5.751868e+01 5.198651e+01
## BID 31.2309384 2.930333e+01 1.290278e+02 3.056954e+01 2.366372e+01
## BLVRB 30.0370017 4.989041e+01 5.371063e+02 4.000646e+01 4.341394e+01
## BSG 59.3419535 7.896545e+01 7.252898e+01 9.359491e+01 6.638857e+01
## BST2 454.4431777 5.488371e+02 1.020885e+03 7.145406e+02 5.555329e+02
## C15orf39 4.1534698 4.962387e+00 2.369471e+01 5.612188e+00 3.202021e+00
## C15orf48 0.0000000 0.000000e+00 4.570036e+00 0.000000e+00 0.000000e+00
## C16orf74 16.9073271 2.284469e+01 1.982149e+01 4.607692e+00 1.824262e+02
## C17orf49 43.3421156 4.839472e+01 3.951806e+01 5.578473e+01 3.412298e+01
## C19orf33 0.0000000 7.926373e+00 0.000000e+00 0.000000e+00 6.655544e+00
## C19orf38 3.8852708 5.057693e+00 2.627770e+02 0.000000e+00 3.005825e+01
## C5AR1 7.6914242 6.919391e+00 1.441184e+02 7.178642e+00 3.680897e+00
## CACTIN 0.9425655 9.705186e-01 8.957417e-01 4.517802e+00 1.208249e+00
## CALM1 219.9718405 3.249561e+02 1.133395e+02 3.451822e+02 1.417586e+02
## CALM3 113.5940115 1.127273e+02 1.020626e+02 1.051918e+02 5.259567e+01
## CALR 104.4668009 1.425749e+02 1.006872e+02 2.184415e+02 1.372829e+02
## CAPNS1 28.3285080 3.671223e+01 9.101543e+01 2.992354e+01 1.754084e+01
## CARHSP1 39.1519099 2.415401e+01 2.636347e+01 4.252122e+01 3.724844e+01
## CARS2 4.5264302 5.391805e+00 2.772122e+01 2.808103e+00 9.430156e+00
## CBFA2T3 0.8440499 2.942975e-01 2.316661e+00 0.000000e+00 8.637592e+00
## CBR1 33.6926690 4.453534e+01 8.918215e+01 3.673470e+01 2.572128e+01
## CCL3 29.9090856 3.027999e+01 9.904089e+02 1.940366e+02 3.056788e+01
## CCL4 19.6175242 1.744920e+01 6.022647e+01 5.932357e+02 3.977257e+01
## CCL5 199.6847550 1.022621e+03 1.277113e+02 1.019931e+04 2.039076e+02
## CCR7 382.3869215 1.367322e+02 9.150186e+00 3.847243e+01 1.243139e+02
## CD19 1.2002304 3.064659e+00 2.791480e+00 5.798455e+00 1.532104e+02
## CD22 6.5552176 3.932897e+00 2.691501e+00 3.295263e+00 6.137696e+01
## CD2BP2 22.8271247 3.038745e+01 3.531081e+01 4.185607e+01 3.544113e+01
## CD300A 12.7140644 2.810323e+01 7.563826e+01 8.562666e+01 7.148143e+00
## CD300C 2.2460930 3.787857e+00 8.646639e+01 4.603830e+00 1.217407e+01
## CD300E 1.2551350 2.756270e+00 6.242355e+01 7.877717e+00 0.000000e+00
## CD300LB 0.6799905 0.000000e+00 1.579298e+01 0.000000e+00 0.000000e+00
## CD300LF 2.9287780 7.155135e+00 8.045634e+01 2.801306e+00 5.346847e+00
## CD320 43.7225546 4.048906e+01 1.826889e+00 8.122302e+01 2.934399e+01
## CD33 1.0920051 4.011534e-01 5.917989e+01 2.245364e+00 0.000000e+00
## CD37 446.0210480 4.748066e+02 5.369561e+02 3.598882e+02 2.097624e+03
## CD40 2.7427645 2.062749e+00 7.396701e+00 2.270071e+00 9.522383e+01
## CD68 19.2588764 1.973362e+01 6.877583e+02 2.673893e+01 3.693306e+01
## CD7 520.9205442 3.439989e+02 2.025774e+01 4.711041e+02 2.857087e+01
## CD79A 43.3365726 5.181972e+01 3.026841e+01 2.658028e+01 4.816654e+03
## CD79B 47.8249569 7.283436e+01 4.097181e+01 9.969250e+01 2.502706e+03
## CDIP1 8.6393733 8.726229e+00 1.090756e+01 1.041814e+01 8.015095e+00
## CDKN2D 140.2441673 1.946742e+02 1.639391e+02 2.989844e+02 2.133099e+02
## CEACAM3 1.1404343 0.000000e+00 4.094264e+00 1.122666e+00 1.260880e+00
## CEBPA 3.3764165 1.582860e+00 3.310392e+01 1.507252e+00 0.000000e+00
## CEBPB 185.9805111 1.528016e+02 7.394999e+02 1.926939e+02 1.100632e+02
## CEBPG 14.4789080 1.708689e+01 3.449475e+01 1.830004e+01 7.494335e+00
## CENPN 1.5032729 2.616917e+00 3.196524e+00 3.368153e+00 1.755446e+00
## CERK 12.8111678 1.394016e+01 1.123336e+01 2.444474e+01 2.782305e+00
## CFD 33.5276672 1.733713e+01 1.133271e+03 3.568599e+01 1.482071e+01
## CHAF1A 1.4658529 2.613819e+00 8.972667e+00 2.453971e+00 3.219064e+00
## CHCHD10 301.0572629 2.825835e+02 3.040202e+02 2.740628e+02 5.236517e+02
## CHEK2 1.8092142 1.752359e+00 5.904992e-01 4.580946e-01 7.157498e+00
## CHMP1B 130.3000870 1.072433e+02 1.587869e+02 8.703196e+01 1.161898e+02
## CHMP4B 87.4644721 9.554961e+01 1.912690e+02 1.190931e+02 1.458983e+02
## CHMP6 25.1294948 2.774256e+01 3.441557e+01 6.549206e+01 2.598680e+01
## CIITA 0.6788259 7.233325e-01 1.753072e+00 8.351624e-01 5.547056e+00
## CINP 12.3168380 1.203939e+01 2.312781e+01 1.180655e+01 9.446602e+00
## CISD3 37.4950329 4.478305e+01 7.091844e+01 8.892808e+01 1.783270e+01
## CKB 3.5160842 1.359477e+00 6.820094e-01 3.938382e+00 1.867767e+00
## CLDN5 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## CLEC10A 0.7273438 1.404887e+00 3.978828e+01 0.000000e+00 0.000000e+00
## CLPP 54.7207889 7.061570e+01 3.154710e+01 6.396010e+01 1.884958e+01
## CMTM3 46.2389148 1.049210e+02 7.189131e+01 9.537650e+01 1.032752e+01
## CMTM5 1.8135363 1.423083e+00 0.000000e+00 1.252322e+00 0.000000e+00
## COMT 13.5417981 2.090584e+01 4.877397e+01 1.834090e+01 1.360174e+01
## COPE 178.9688340 2.265235e+02 2.925149e+02 2.869030e+02 2.007844e+02
## CORO1A 444.5494917 5.394662e+02 4.460411e+02 5.988750e+02 4.637858e+02
## COTL1 124.8737667 2.343218e+02 8.464099e+02 1.894742e+02 1.657150e+02
## COX5A 290.0735312 3.733611e+02 5.378810e+02 4.696730e+02 3.632267e+02
## COX6B1 673.3079901 7.714533e+02 1.020420e+03 6.682147e+02 6.070488e+02
## CPPED1 4.8156724 5.807048e+00 5.207239e+01 4.721149e+00 3.676411e+00
## CRELD2 12.2959423 2.627252e+01 1.335236e+01 7.096324e+01 2.031315e+01
## CRIP1 196.9931663 3.825106e+02 3.145246e+02 3.006794e+02 1.618048e+02
## CRIP2 27.2707845 6.274355e+01 1.874909e+00 1.309262e+01 1.315286e+01
## CSK 42.4841072 5.311290e+01 5.919544e+01 7.049768e+01 8.646305e+01
## CST3 69.4976423 7.439540e+01 3.514783e+03 8.562660e+01 8.595753e+01
## CST7 47.9051199 1.921424e+02 3.095789e+01 3.310233e+03 7.266010e+01
## CSTB 157.4690322 1.869631e+02 3.457918e+02 2.246439e+02 8.328671e+01
## CTNNBL1 33.1551985 2.628872e+01 3.852725e+01 4.031703e+01 3.401584e+01
## CTSA 17.0968579 2.670598e+01 2.426608e+01 1.993189e+01 8.527548e+00
## CTSH 2.0877579 6.130248e+00 5.124252e+01 7.415362e+00 2.169684e+01
## CTSZ 0.3486078 1.552684e+00 2.183755e+01 2.157761e+00 6.896403e+00
## CXCL16 6.5738379 2.349792e+00 2.896780e+01 2.782554e+00 1.004024e+01
## CYB5B 8.5933958 2.637652e+01 2.585356e+00 1.421128e+01 1.262394e+01
## CYBA 362.7730619 5.235995e+02 1.722263e+03 1.014396e+03 1.023142e+03
## CYLD 15.8067260 2.167677e+01 3.107261e+00 7.639091e+00 1.102221e+01
## CYTH4 26.0400868 2.418589e+01 2.900948e+01 3.162583e+01 1.904432e+01
## DAAM1 1.9681682 3.148796e+00 1.311390e-01 9.465483e-01 1.955552e+00
## DDX5 357.1969387 3.543474e+02 1.056193e+02 3.680754e+02 3.282997e+02
## DEDD2 46.5666278 5.672157e+01 4.232805e+01 7.242684e+01 4.894897e+01
## DENND6B 1.4477371 2.866126e+00 1.788345e+01 3.577981e+00 5.548967e-01
## DESI1 13.6605851 2.527914e+01 1.780035e+01 1.480840e+01 1.158989e+01
## DET1 2.0090447 8.537806e-01 1.649885e+00 5.900534e-01 4.804973e-01
## DHRS4L2 37.5762007 4.857169e+01 1.188653e+02 4.173904e+01 5.075030e+01
## DHRS7 109.2756869 1.810136e+02 1.265116e+02 2.230103e+02 8.341668e+01
## DNAJB1 94.6941801 8.731356e+01 1.866804e+01 9.632908e+01 2.292405e+01
## DNASE2 24.2617436 2.461288e+01 2.354054e+01 2.346439e+01 1.758831e+01
## DPEP2 25.9115358 2.270068e+01 3.668449e+01 1.227505e+01 3.025268e+01
## DSTN 71.0556052 9.781881e+01 2.243800e+01 1.083830e+02 3.278464e+01
## DTD2 6.5349414 7.130291e+00 4.209321e+00 1.085222e+01 6.643338e+00
## DUSP3 1.2805083 2.266421e+00 1.128004e+01 3.075951e+00 3.985007e+00
## EGLN3 0.9098907 2.156882e+00 0.000000e+00 4.595290e-01 0.000000e+00
## EIF3G 312.2956050 3.352572e+02 2.576965e+02 2.960292e+02 2.656189e+02
## EIF4A1 213.2461935 3.174661e+02 4.948613e+02 3.172249e+02 2.116580e+02
## EIF6 126.2297182 1.366922e+02 1.975209e+02 2.137309e+02 2.270904e+02
## EMC7 104.0504637 1.813569e+02 1.702682e+02 1.773545e+02 1.198851e+02
## EMC8 22.6055629 1.341822e+01 2.399624e+01 1.792198e+01 1.908731e+01
## EMP3 757.9683847 1.424060e+03 1.590760e+03 1.396605e+03 8.732780e+02
## EPN1 1.8629141 2.643211e+00 4.284164e+00 2.874547e+00 3.178381e+00
## ERCC1 17.7069251 2.007850e+01 3.371595e+01 1.903461e+01 2.524310e+01
## ETFA 10.9650101 1.583156e+01 1.469769e+01 1.396926e+01 1.803958e+01
## ETHE1 78.2271251 1.747034e+02 1.451336e+02 1.551708e+02 2.823213e+01
## EVI2B 228.2045221 2.493481e+02 2.684017e+02 1.557320e+02 3.586637e+02
## EVL 145.7246988 1.586666e+02 2.172643e+01 1.227576e+02 8.755575e+01
## FAH 0.9352185 1.252074e+00 2.189248e+00 1.009074e+00 0.000000e+00
## FAM110A 18.6851561 1.913401e+01 3.503252e+01 2.114300e+01 1.204604e+01
## FAM118A 5.3019230 6.265161e+00 3.427401e+00 3.875863e+00 6.306094e+00
## FAM32A 83.2749877 9.455863e+01 1.509920e+02 8.165131e+01 9.650780e+01
## FCER2 0.8036517 4.285682e+00 6.312531e+00 7.842462e+00 4.028635e+02
## FCGRT 39.9678328 2.703880e+01 3.169489e+02 2.383299e+01 4.059798e+01
## FCHO1 3.9079245 4.637047e+00 4.526792e+00 3.681094e+00 1.036916e+01
## FES 4.1589362 4.815568e-01 4.008066e+01 6.208293e-01 1.931035e+00
## FKBP1A 90.5824065 1.048052e+02 2.485942e+02 8.250438e+01 1.053066e+02
## FKBP3 92.7893260 8.240152e+01 2.079375e+01 5.678306e+01 6.938136e+01
## FLT3LG 166.4635031 1.666689e+02 1.684932e+01 1.322653e+02 2.196150e+01
## FMNL1 41.3510100 4.160468e+01 3.846643e+01 4.486241e+01 4.228321e+01
## FN3K 2.9264772 3.564055e+00 2.229085e+00 1.323400e+00 0.000000e+00
## FNTB 1.2768399 1.712019e+00 1.284693e+00 1.002799e+00 2.225458e+00
## FOSB 139.4051215 1.455180e+02 2.789623e+02 1.288333e+02 1.566481e+02
## FPR1 7.1463009 7.696814e+00 2.602059e+02 4.021688e+00 2.260348e+00
## FXYD5 723.1331356 9.961044e+02 4.590185e+02 5.710951e+02 5.810497e+02
## G6PC3 2.6773008 2.037147e+00 4.261296e+00 2.593292e+00 5.458460e+00
## GAA 0.2810469 4.109833e+00 1.189303e+01 5.984568e+00 2.028969e+00
## GABARAP 253.4620373 2.793812e+02 1.325088e+03 2.739133e+02 3.858084e+02
## GAS2L1 0.9332817 9.843250e-01 8.772092e+00 0.000000e+00 1.707189e+00
## GAS6 0.3799665 0.000000e+00 1.224468e+00 0.000000e+00 0.000000e+00
## GAS7 0.0000000 9.131180e-01 1.337946e+01 1.718354e+00 2.541881e+00
## GCH1 28.1572198 2.851090e+01 2.743105e+01 2.597939e+01 2.709483e+01
## GCHFR 83.2513385 1.076684e+02 7.114255e+01 1.359453e+02 3.465000e+01
## GLOD4 38.0877886 6.146498e+01 1.244680e+01 4.019917e+01 3.701455e+01
## GMFG 1113.6457441 1.373498e+03 1.291477e+03 1.394474e+03 8.309434e+02
## GMIP 11.2133943 1.659371e+01 1.435179e+01 1.326756e+01 1.223593e+01
## GNA15 1.4060302 5.985552e+00 2.498036e+01 2.380471e+00 1.301743e+00
## GNAS 13.6202798 1.634802e+01 2.393292e+01 1.838806e+01 1.961351e+01
## GNB5 4.5951339 3.572034e+00 1.027008e+00 3.335408e+00 6.254667e+00
## GNG2 22.0383966 2.711073e+01 1.122033e+01 3.149297e+01 1.780591e+01
## GNG7 5.1147584 3.802063e+00 7.775763e+00 1.944693e+00 1.217452e+02
## GNGT2 1.8595954 1.797580e+00 1.876591e+01 1.308901e+01 4.562089e+00
## GP1BA 2.8432765 1.513606e+00 5.052792e-01 1.191549e+00 0.000000e+00
## GP6 0.0000000 0.000000e+00 9.434357e-01 1.561552e+00 0.000000e+00
## GPR18 23.8046247 1.626149e+01 5.385440e+00 3.819517e+01 1.276056e+02
## GPR183 131.3321119 1.977619e+02 2.415395e+01 2.044528e+02 1.505092e+02
## GPX4 527.4599754 4.955734e+02 8.191886e+02 4.874822e+02 4.365855e+02
## GRAP2 33.8965427 3.527659e+01 5.121659e+00 4.137976e+01 1.372285e+01
## GRB2 31.7209833 4.220983e+01 7.469874e+01 3.255036e+01 1.027359e+02
## GRN 21.1735125 2.544978e+01 4.879151e+02 2.776263e+01 4.881160e+01
## GUCD1 7.5573009 5.782669e+00 1.073044e+01 9.084720e+00 6.628807e+01
## GZMB 26.1173083 3.015634e+01 3.198761e+01 8.339585e+02 2.245301e+01
## GZMH 22.0895281 2.170556e+01 2.318893e+01 2.490943e+03 3.289562e+01
## GZMM 412.4028284 3.752728e+02 2.904537e+01 1.627811e+03 3.281175e+01
## HAUS1 38.8168800 3.423928e+01 2.804518e+01 6.183892e+01 6.089448e+01
## HCK 10.0600860 8.447296e+00 1.581988e+02 1.296025e+01 4.326645e+01
## HCST 1883.2358044 2.228376e+03 1.782126e+03 4.603130e+03 3.326430e+02
## HERPUD1 33.0403271 5.699129e+01 5.768985e+01 5.578496e+01 1.019009e+02
## HEXA 5.5924772 5.388870e+00 8.524704e+00 8.384898e+00 1.112298e+01
## HEXIM2 5.0593371 9.649575e+00 8.647591e+00 6.992696e+00 1.745583e+00
## HMG20B 14.2724780 2.906087e+01 1.288466e+01 2.547484e+01 1.764312e+01
## HMGN1 249.6067039 2.458486e+02 8.900641e+01 1.952844e+02 2.994773e+02
## HMOX1 3.9722488 5.897879e+00 6.933617e+01 5.261992e+00 5.621555e+00
## HMOX2 93.4601152 8.961104e+01 1.978440e+01 1.024189e+02 4.644612e+01
## HSBP1 7.9466192 1.402403e+01 3.743618e+01 1.363827e+01 1.253352e+01
## HSCB 51.9293560 2.808559e+01 3.919037e+01 2.301616e+01 5.849851e+01
## HSH2D 23.3673121 1.600107e+01 2.320748e+01 2.744390e+01 5.304167e+01
## ICAM2 104.9849946 1.522262e+02 1.000417e+02 1.100285e+02 8.105871e+01
## ICAM3 213.6545570 3.131475e+02 2.079233e+02 2.648055e+02 2.252532e+02
## ICAM4 0.0000000 0.000000e+00 6.011100e+00 0.000000e+00 0.000000e+00
## ID1 27.3324197 2.753439e+01 2.542769e+02 5.059161e+01 1.686461e+01
## IDH2 81.7987800 6.642770e+01 1.057652e+02 1.288546e+02 5.490147e+01
## IDH3A 6.3384121 6.660201e+00 5.107074e+00 9.353586e+00 1.257400e+01
## IER2 514.7528803 7.913080e+02 5.576318e+02 7.393107e+02 4.800000e+02
## IFI27L2 94.9863655 1.104885e+02 2.087690e+02 1.582898e+02 1.565034e+02
## IFI30 0.8711953 1.965298e+00 3.746473e+02 1.394304e+00 2.642857e+01
## IFI35 65.6851805 1.406794e+02 2.163867e+02 1.002621e+02 1.431716e+02
## IFNGR2 29.3288995 3.182094e+01 2.093481e+02 9.294883e+00 1.230427e+02
## IGFLR1 28.5375689 3.148389e+01 5.023927e+01 3.582094e+01 5.015025e+01
## IGLL5 7.8354675 7.324477e+00 3.731330e+00 7.383144e+00 2.235888e+03
## IGSF6 1.7653616 2.957987e+00 1.703283e+02 3.855653e+00 4.530380e+00
## IL10RB 12.5094674 1.360118e+01 2.180924e+01 1.230667e+01 1.727057e+01
## IL12RB1 15.3117551 3.103201e+01 1.222875e+01 3.690027e+01 8.036048e+00
## IL17RA 2.6812464 2.481769e+00 1.816770e+01 2.434913e+00 1.131749e+00
## IL2RB 5.4126660 8.600203e+00 2.013117e+00 3.347719e+01 2.021877e+00
## IL32 832.3363586 1.625250e+03 4.042575e+01 1.861302e+03 6.055256e+01
## IL4R 12.4448473 1.228049e+01 5.222385e+00 6.980029e+00 5.697364e+01
## IMPA2 2.2155534 3.950815e+00 2.556646e+01 7.170921e-01 3.168131e+00
## IQGAP1 4.6307875 9.554139e+00 1.873075e+01 1.022462e+01 1.270004e+01
## IRF8 4.6652004 3.643944e+00 2.065761e+01 2.857416e+00 1.246738e+02
## ISG20 127.6274712 1.774867e+02 2.326894e+01 1.479935e+02 3.240464e+02
## ISOC2 12.4079285 1.754966e+01 3.090114e+01 1.591207e+01 1.758670e+01
## ITGA2B 0.7877406 0.000000e+00 2.444406e-01 0.000000e+00 0.000000e+00
## ITGAL 7.1767479 8.289571e+00 1.150235e+01 2.133941e+01 5.254328e+00
## ITGAM 0.0000000 1.645938e+00 2.430168e+01 6.283364e+00 5.884002e+00
## ITGAX 0.6318757 1.060033e+00 9.258896e+00 4.932007e-01 3.414492e+00
## ITGB2 68.0875316 9.152198e+01 1.597227e+02 1.588881e+02 4.024714e+01
## ITGB3 0.3630808 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## ITPK1 1.4784948 3.371552e+00 1.088213e+01 3.892340e+00 3.686027e+00
## JOSD2 46.8275103 8.496055e+01 1.423153e+02 9.033980e+01 2.392190e+01
## JUND 84.5327259 6.969539e+01 3.344933e+02 7.380389e+01 1.085701e+02
## KIAA0513 2.1118946 4.946413e-01 5.312287e+00 3.833993e-01 5.673093e-01
## KIAA0930 1.6362173 1.015407e+00 1.339044e+01 2.790273e+00 5.261622e+00
## KIF16B 0.8794821 1.374169e+00 2.637058e+00 1.916759e+00 4.779050e-01
## KIFC3 0.1832536 0.000000e+00 1.689330e-01 2.046465e+00 0.000000e+00
## KIR2DL3 0.0000000 3.012111e+00 2.037807e+00 7.317479e+00 0.000000e+00
## KIR3DL2 0.0000000 9.784920e+00 2.018670e+00 1.693896e+01 0.000000e+00
## KPNA2 2.9964222 7.830832e+00 4.209595e+00 6.775478e+00 7.887960e+00
## KSR1 0.1999794 6.218049e-01 2.204728e+00 1.276273e+00 1.049436e+00
## LACTB 12.3743085 7.535257e+00 4.379762e+01 9.916963e+00 2.631925e+01
## LAIR1 8.3315751 6.205316e+00 2.293865e+01 1.486715e+01 1.834960e+01
## LAMP1 45.3542891 4.698720e+01 3.118018e+01 6.001092e+01 4.218502e+01
## LASP1 14.9356260 1.833914e+01 1.211166e+01 2.608534e+01 7.512593e+00
## LAT 114.4566301 1.301464e+02 5.227280e+00 1.380617e+02 1.004470e+01
## LDLRAD4 1.1450190 5.948329e-01 8.380827e-01 8.668177e-01 6.662176e-01
## LGALS1 262.6791926 9.903369e+02 7.929078e+03 1.075933e+03 2.484334e+02
## LGALS2 57.2558054 4.418881e+01 4.807282e+03 2.405202e+01 6.597949e+01
## LGALS3 27.4129725 1.158321e+02 8.444795e+02 1.133774e+02 3.129205e+01
## LGALS9 15.1509455 2.167302e+01 8.223794e+01 1.594767e+01 3.379573e+01
## LGMN 4.8695049 2.863669e+00 2.800230e+00 2.102061e+00 4.296718e+00
## LILRA1 4.9083228 4.264212e+00 2.348206e+01 2.585267e+00 3.077850e+00
## LILRA2 0.3476922 5.689347e-01 2.313382e+01 2.314335e+00 4.783845e+00
## LILRA4 0.8390216 0.000000e+00 6.877201e-01 3.550973e+00 4.745069e+00
## LILRA5 4.4376019 3.701223e+00 2.836786e+02 7.985517e+00 8.705679e+00
## LILRB1 2.2510928 1.821679e+00 1.728326e+01 1.641648e+00 1.729578e+01
## LILRB2 7.2049691 8.700080e+00 7.074509e+01 4.675946e+00 5.693688e+00
## LILRB3 1.7712835 2.609690e+00 3.672151e+01 3.777072e-01 2.490670e+00
## LILRB4 1.0168485 6.771534e-01 2.733827e+01 1.144142e+00 2.119996e+00
## LIMD2 406.4854293 3.715667e+02 1.797165e+02 2.926482e+02 5.387046e+02
## LINC00528 12.2398254 5.151756e+00 5.232693e+00 3.459315e+00 4.354205e+00
## LINC00926 1.8114136 4.995212e+00 3.080322e+00 2.159627e+00 5.104402e+02
## LITAF 97.3195326 1.000953e+02 4.002595e+01 2.048326e+02 7.779189e+01
## LRRC25 3.2736842 5.967976e+00 1.247389e+02 7.996546e+00 1.011236e+01
## LYL1 12.1474496 1.133896e+01 8.383785e+01 6.222206e+00 1.318939e+02
## LYSMD2 93.0443173 1.221731e+02 1.425466e+02 1.251319e+02 1.474973e+02
## MAFB 4.2544181 2.548619e+00 1.382574e+02 2.947797e+00 2.457769e+00
## MAFG 1.6960696 2.066238e+00 9.623337e+00 1.236023e+00 6.231838e+00
## MAP2K3 24.5130189 3.863898e+01 5.117656e+01 6.120526e+01 4.703311e+01
## MAP3K7CL 3.1901506 1.512318e+00 6.703418e+00 5.429673e+00 3.862518e+00
## MAP4K1 16.7842524 2.996672e+01 6.747701e+00 3.970211e+01 3.027337e+01
## MAPK1 10.6860592 1.774723e+01 1.650246e+01 2.457953e+01 1.873917e+01
## MATK 5.6238445 7.687965e+00 2.959586e+00 1.013751e+02 4.634143e+00
## MAX 30.2384104 3.418942e+01 3.034537e+01 3.295212e+01 3.582012e+01
## MBD2 0.8970136 2.994289e+00 7.586875e+00 3.716467e+00 3.585518e+00
## MBOAT7 6.4783065 8.631010e+00 2.806080e+01 6.030332e+00 2.859248e+00
## MBP 6.0748991 7.855089e+00 4.289400e+00 1.065219e+01 2.779943e+00
## METTL9 38.8796149 5.582682e+01 6.857917e+01 6.644592e+01 5.107361e+01
## MIDN 12.7298779 1.445091e+01 5.389090e+01 3.205148e+01 1.826213e+01
## MIEN1 69.1534430 1.122866e+02 8.402302e+01 1.482783e+02 1.150043e+02
## MIF 1014.7508381 1.142510e+03 4.942821e+02 9.139278e+02 8.468008e+02
## MILR1 6.1440657 1.118435e+00 2.643563e+01 0.000000e+00 3.008364e+01
## MIR142 1688.2857479 1.766713e+03 3.083425e+03 4.008401e+03 3.283143e+03
## MIR24-2 741.5365453 1.113929e+03 4.910830e+03 1.444486e+03 1.039274e+03
## MIS18BP1 8.1202346 7.083814e+00 1.081032e+01 1.079817e+01 4.366558e+01
## MLH3 5.2170888 2.574884e+00 7.141892e-01 5.475438e+00 2.268829e+00
## MLX 34.3097132 2.795056e+01 3.662318e+01 4.983868e+01 2.289738e+01
## MMD 6.9640494 6.484859e+00 1.504224e+00 1.024124e+01 1.088595e+01
## MOB3A 18.3211970 1.756060e+01 4.501792e+01 2.610142e+01 2.885223e+01
## MPST 4.9514106 4.456915e+00 2.238095e+01 1.109061e+01 3.404768e+00
## MRPL27 33.1962988 3.437236e+01 5.285857e+01 3.428323e+01 3.677727e+01
## MRPL54 269.3853121 2.841884e+02 3.818013e+02 2.806554e+02 2.627049e+02
## MRPS6 57.7057199 5.198480e+01 3.934269e+01 7.418636e+01 4.558278e+01
## MSRB1 17.6424435 2.457926e+01 1.919652e+02 3.782570e+01 1.955751e+01
## MT-ND3 784.7775885 7.106521e+02 1.060300e+03 9.656561e+02 6.945920e+02
## MT2A 352.8267103 6.432866e+02 1.032190e+03 8.681286e+02 7.378819e+01
## MTHFS 45.5190671 4.485797e+01 6.588588e+01 3.719463e+01 5.248601e+01
## MVD 9.3910347 9.196455e+00 1.182305e+01 2.340358e+01 1.516650e+01
## MX2 8.2357563 9.528834e+00 1.187959e+01 1.127326e+01 5.288772e+00
## MYADM 59.1056279 7.506696e+01 1.205951e+02 4.223428e+01 4.226083e+01
## MYL12A 1214.5200800 1.604559e+03 4.486664e+02 1.617621e+03 8.956332e+02
## MYL12B 885.4176751 1.197508e+03 6.151561e+02 1.329712e+03 6.731071e+02
## MYL9 1.7335478 1.243851e+00 2.650883e+00 2.429621e+00 3.458738e+00
## MYO1F 10.8012585 3.238155e+01 4.985188e+01 7.785999e+01 9.507355e+00
## MYO9B 4.8847403 4.869478e+00 3.718777e+01 4.904057e+00 5.317693e+00
## NAGA 4.2326407 7.037106e+00 2.649015e+01 1.788642e+00 6.158470e+00
## NAPA 22.3511887 2.437311e+01 3.018358e+01 3.522245e+01 1.675452e+01
## NCF4 4.1821516 1.878233e+01 8.031701e+01 2.080602e+00 8.187402e+01
## NDRG2 4.2726013 5.680429e+00 1.032298e+00 8.271016e-01 1.579677e+00
## NDUFA11 125.2129280 1.271340e+02 1.990093e+02 1.281515e+02 1.004221e+02
## NDUFA13 224.2686454 2.643409e+02 3.472991e+02 2.588648e+02 2.222305e+02
## NDUFB1 89.9246021 8.060522e+01 1.612521e+02 9.355035e+01 5.311696e+01
## NDUFB10 447.7493626 4.603424e+02 6.229545e+02 5.522567e+02 4.150461e+02
## NDUFB7 494.5109734 6.365066e+02 8.221574e+02 9.587319e+02 6.308311e+02
## NDUFS7 44.0901751 4.071999e+01 9.112166e+01 4.486073e+01 4.086141e+01
## NDUFV3 10.7926601 9.478673e+00 2.334076e+01 9.022977e+00 6.703381e+00
## NFKBIA 187.3829794 3.065349e+02 6.064846e+02 3.586298e+02 2.265153e+02
## NKG7 149.0918473 2.646173e+02 2.646920e+02 1.319077e+04 1.926235e+02
## NLK 2.3987893 2.698808e+00 3.444270e-01 2.668019e+00 3.286796e+00
## NME1 18.5481060 2.230713e+01 2.395560e+01 2.043439e+01 3.099222e+01
## NME1-NME2 68.0973243 4.840695e+01 7.184954e+01 4.785502e+01 2.605547e+01
## NOP10 171.2227481 2.182368e+02 4.893059e+02 2.300245e+02 1.729598e+02
## NOSIP 402.9423994 3.085917e+02 6.368417e+01 2.039007e+02 5.243976e+01
## NPC2 133.9393982 1.460425e+02 1.156566e+03 1.060692e+02 2.383801e+02
## NT5C 60.4086418 1.183331e+02 6.706111e+01 1.959821e+02 2.573243e+02
## NT5M 1.6680341 0.000000e+00 2.061149e+00 1.811632e+00 0.000000e+00
## OAZ1 754.7016235 9.012698e+02 2.745009e+03 1.051761e+03 1.285049e+03
## OAZ2 15.3402426 1.287036e+01 5.567440e+01 1.563380e+01 2.715294e+01
## OGFOD3 9.5184502 1.003731e+01 8.243651e+00 7.222171e+00 6.474428e+00
## OLIG1 0.0000000 0.000000e+00 2.614159e+01 0.000000e+00 0.000000e+00
## OSCAR 2.1360617 4.229825e+00 9.888658e+01 6.820272e-01 8.128596e-01
## OXA1L 55.2570452 5.368641e+01 7.017237e+01 3.896737e+01 7.014937e+01
## P2RX1 2.3114093 1.125968e+00 8.986436e+00 8.918560e-01 4.386063e+00
## P2RX5 6.6181780 5.153733e+00 2.892537e+00 9.370402e+00 1.635215e+02
## P4HB 15.6224378 3.353474e+01 3.083282e+01 4.530233e+01 2.006223e+01
## PACSIN2 6.9326795 1.187962e+01 1.516751e+01 1.305246e+01 3.137998e+01
## PARVB 4.0620130 4.958875e+00 1.541230e+01 7.683114e+00 7.122940e+00
## PARVG 15.6596316 1.583483e+01 3.566633e+01 1.536776e+01 2.792533e+01
## PATL2 7.5380562 1.239570e+01 1.332787e+00 3.381208e+01 1.941618e+01
## PCK2 5.0676702 5.865155e+00 6.186050e+00 6.545283e+00 1.105743e+01
## PCNA 114.1775651 6.460188e+01 1.252526e+01 8.309558e+01 3.870875e+01
## PCP2 0.0000000 9.906072e+00 0.000000e+00 0.000000e+00 2.335050e+01
## PDCD5 120.0420523 1.820143e+02 5.627527e+01 1.181668e+02 8.527927e+01
## PDIA3 31.4534682 5.985712e+01 2.337785e+01 7.526637e+01 3.479622e+01
## PDXK 2.5353051 4.356910e+00 1.512626e+01 3.053815e+00 3.910324e+00
## PER1 4.7044997 4.894429e+00 1.592906e+01 6.984852e+00 3.673490e+00
## PFKL 11.6673773 1.458990e+01 1.342112e+01 9.943141e+00 2.069007e+01
## PGLS 127.8354779 8.743036e+01 2.909635e+02 1.081078e+02 9.262875e+01
## PIK3IP1 216.9246362 1.506496e+02 1.343865e+01 8.899170e+01 5.755567e+01
## PITPNC1 13.3835072 1.453652e+01 4.594730e+00 2.265822e+01 3.327090e+00
## PKIG 5.4643137 3.923879e+00 1.863394e+00 0.000000e+00 2.326378e+02
## PKM 69.8210860 1.028247e+02 1.082738e+02 7.270895e+01 4.604369e+01
## PKN1 32.7490353 2.538244e+01 3.359726e+01 3.194836e+01 2.705229e+01
## PLA2G4C 0.0000000 3.952990e-01 4.988603e-01 6.133664e-01 0.000000e+00
## PLAGL2 0.8844910 1.185877e+00 6.419254e+00 1.282930e+00 7.676278e-01
## PLAUR 6.3380282 9.157756e+00 1.050184e+02 2.645832e+00 8.488857e+00
## PLD4 1.1728788 2.037703e+00 1.486335e+01 5.949659e+00 7.991857e+01
## PLEKHF1 9.3527742 2.387061e+01 4.047549e+00 7.138271e+01 1.614011e+01
## PLIN3 10.0844696 2.340884e+01 6.183468e+01 1.329377e+01 1.444245e+01
## PLXNB2 3.1013249 9.720825e-01 1.256984e+01 0.000000e+00 6.179326e-01
## PMAIP1 37.3189599 1.211119e+02 6.399787e+01 1.132579e+02 1.464606e+02
## PNMA1 14.6337785 3.324079e+01 4.502793e+00 4.733371e+00 1.050032e+01
## POLR3K 45.3308527 7.978430e+01 5.724214e+01 1.377157e+02 9.620957e+01
## POU2F2 6.0225565 1.221919e+01 4.244373e+01 6.324382e+00 5.034391e+01
## PPCDC 6.5551926 7.520390e+00 1.399398e+01 9.102803e+00 3.736025e+00
## PPDPF 2177.4914393 2.546765e+03 1.717105e+03 2.299959e+03 2.310986e+03
## PPIB 164.0249606 2.564085e+02 1.694717e+02 3.118998e+02 1.600266e+02
## PPM1F 2.0932795 8.818344e-01 9.138377e+00 2.099321e+00 1.145496e+00
## PPM1N 11.3869183 7.061330e+00 1.043467e+00 1.666579e+01 9.396234e+00
## PPP1R14A 4.6014780 0.000000e+00 4.157033e+00 2.849138e+00 2.795090e+02
## PPP2R3C 25.8132606 2.019595e+01 1.290538e+01 6.134635e+01 2.209450e+01
## PPP2R5C 25.0997974 5.159260e+01 6.601530e+00 4.793191e+01 1.672468e+01
## PPP4C 109.2296694 1.348570e+02 1.909345e+02 1.504816e+02 1.170697e+02
## PRAM1 5.1463987 3.414934e+00 7.393357e+01 1.827557e+00 4.972104e+00
## PRDX2 306.4428242 3.729828e+02 5.516032e+01 2.975359e+02 1.198342e+02
## PRKCB 9.2513800 1.011598e+01 1.655301e+01 1.810743e+01 5.284990e+01
## PRKCH 24.8221151 2.388444e+01 2.317345e+00 3.941582e+01 5.842259e+00
## PRMT2 51.1588509 4.019170e+01 2.464809e+01 8.083659e+01 1.747333e+01
## PRR14L 2.4060487 4.247613e-01 1.575849e+00 2.767591e-01 7.414119e-01
## PRR5 10.8226735 1.445145e+01 2.553214e+00 8.028803e+01 6.659155e+00
## PSENEN 85.0366903 1.158983e+02 1.404098e+02 1.614688e+02 1.000331e+02
## PSMA4 16.9713164 2.478496e+01 2.932814e+01 2.249823e+01 2.184525e+01
## PSMA7 517.7355554 5.712836e+02 8.115585e+02 7.003229e+02 4.745981e+02
## PSMB10 267.2924824 3.469889e+02 3.002476e+02 3.490390e+02 2.771057e+02
## PSMB3 315.0145522 4.124549e+02 6.346046e+02 4.582959e+02 4.606229e+02
## PSMC4 70.3035250 9.200734e+01 3.865065e+01 1.214156e+02 8.745712e+01
## PSMC5 92.9932638 1.560040e+02 6.112464e+01 1.110273e+02 1.023637e+02
## PSMD8 77.8759845 8.977724e+01 7.611575e+01 1.056843e+02 1.064362e+02
## PSME1 1101.5648816 1.181252e+03 8.343687e+02 1.277394e+03 1.048389e+03
## PSME2 231.3891305 3.091645e+02 3.505078e+02 3.237378e+02 2.265581e+02
## PTGDR 5.9853492 1.100514e+01 1.584162e+00 8.255512e+01 4.619763e+00
## PTGER2 49.3160168 1.200485e+02 5.585740e+01 1.376831e+02 1.312788e+01
## PVALB 2.1267125 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## PYCARD 131.8677964 1.644482e+02 1.112005e+03 2.423053e+02 3.288313e+02
## PYGL 2.0407261 1.774001e+00 3.120882e+01 4.374736e+00 4.703721e+00
## RAB11A 30.3554756 2.581507e+01 1.591816e+01 2.742450e+01 2.674853e+01
## RAB22A 4.8886231 5.892972e+00 4.168393e+00 1.040574e+01 4.268625e+00
## RAB27B 0.2300914 1.361863e+00 2.631977e-01 2.506612e+00 7.476173e-01
## RAB31 0.0000000 3.087687e-01 4.091288e+01 7.884898e-01 3.682822e+00
## RAB34 10.8945114 1.671080e+00 4.240952e+01 2.117374e+01 5.694349e+00
## RAB4B 18.1546300 2.197491e+01 1.461974e+01 2.545853e+01 3.817517e+01
## RAB5C 92.4665224 1.397401e+02 1.403254e+02 1.441207e+02 1.450575e+02
## RAB8A 30.9855963 5.839581e+01 6.694265e+01 6.858386e+01 5.088947e+01
## RABGGTA 9.5183470 1.539786e+01 1.645619e+01 1.236810e+01 1.417764e+01
## RAC2 574.4279605 6.572965e+02 3.061422e+02 6.493777e+02 5.635739e+02
## RALY 29.2457278 5.995457e+01 5.148296e+01 5.077181e+01 3.421380e+01
## RASAL3 30.2283511 3.948031e+01 1.003018e+01 5.161180e+01 2.793809e+01
## RASSF2 4.8733823 7.140489e+00 1.552004e+01 2.921323e+00 9.004811e+00
## RBM39 48.0796091 5.245402e+01 2.546664e+01 5.462109e+01 7.801609e+01
## RBM42 43.1359121 4.705185e+01 4.053467e+01 4.571418e+01 2.934019e+01
## RBX1 83.4285181 1.243885e+02 1.545538e+02 1.399890e+02 1.144164e+02
## RCN2 18.0693664 1.999360e+01 5.196767e+00 2.374702e+01 3.330887e+01
## RDH11 18.5190550 1.679035e+01 7.634379e+00 2.207045e+01 2.117300e+01
## RETN 22.8179218 7.010847e+00 4.998004e+02 9.413953e+00 0.000000e+00
## RGL4 53.6884357 5.445523e+01 3.554754e+00 4.082939e+01 6.994435e+00
## RGS19 93.0984873 1.632859e+02 2.170739e+02 2.627816e+02 2.100113e+02
## RIN3 10.5310252 7.707287e+00 1.588798e+01 6.452568e+00 1.365253e+01
## RNASE2 7.9273105 0.000000e+00 1.467904e+02 0.000000e+00 0.000000e+00
## RNASE6 14.9318420 3.366651e+01 4.468098e+02 2.497812e+01 5.171427e+02
## RNF125 15.1745666 1.664566e+01 1.282649e+01 2.395635e+01 4.244059e+00
## RNF126 63.2215997 7.411431e+01 2.588390e+01 9.301717e+01 7.039319e+01
## RNF135 8.2967817 5.371628e+00 4.972963e+01 3.602323e+00 3.928898e+01
## RNF167 66.0229288 1.218848e+02 4.008104e+01 1.371772e+02 6.234817e+01
## RNF213 9.4125164 1.525973e+01 1.087404e+01 1.188440e+01 8.971762e+00
## RORA 10.7943017 1.619742e+01 1.175997e+00 2.434245e+01 2.247548e+00
## RPL36AL 3590.5242629 3.693601e+03 2.123506e+03 3.382445e+03 2.526197e+03
## RPS21 4190.4956943 3.923665e+03 1.586895e+03 2.903348e+03 4.262402e+03
## RRAS 7.4120680 8.859449e+00 4.138998e+01 1.284270e+01 6.464548e+00
## RRP7A 14.1095281 2.186144e+01 2.534027e+01 2.034692e+01 2.943208e+01
## RSL1D1 122.5175824 7.955363e+01 6.301854e+01 7.754429e+01 8.407314e+01
## RTN1 0.0000000 3.142416e-01 2.469104e+00 0.000000e+00 0.000000e+00
## S100B 86.1954041 3.333049e+00 2.173377e+00 1.804471e+01 5.126724e+00
## S1PR5 3.2134675 1.635482e+00 1.158517e+00 8.550692e+01 4.815003e+00
## SAMD14 0.3137688 2.690351e-01 0.000000e+00 0.000000e+00 0.000000e+00
## SAMHD1 23.0255286 2.668219e+01 6.006052e+01 2.623211e+01 5.323458e+00
## SAT2 93.4931075 6.866643e+01 3.398279e+02 1.233025e+02 1.206671e+02
## SCAMP4 8.8093052 4.745325e+00 4.624755e+00 5.545166e+00 6.876382e+00
## SCIMP 5.0535077 5.939608e+00 4.931108e+01 3.540985e+00 2.811855e+01
## SCN1B 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## SCO2 38.4324228 7.576482e+01 3.227628e+02 6.536920e+01 5.535721e+01
## SCPEP1 16.7601188 1.024963e+01 1.203371e+02 2.158682e+01 1.141256e+02
## SDF2L1 121.8390835 1.890896e+02 1.489446e+02 2.744342e+02 9.586492e+01
## SEC11A 59.1752950 7.102540e+01 1.326335e+02 8.298642e+01 8.302952e+01
## SEC14L1 2.2311306 4.171470e+00 5.643168e+00 3.815241e+00 5.633623e+00
## SEC14L5 0.0000000 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## SECTM1 0.9848055 1.273673e+00 2.312623e+01 1.320066e+00 0.000000e+00
## SERF2 224.0577514 2.760326e+02 4.411845e+02 3.480699e+02 2.355624e+02
## SERPINA1 19.8649621 1.900675e+01 3.130202e+02 1.894151e+01 1.990292e+01
## SERPINF1 0.0000000 0.000000e+00 3.370883e+00 0.000000e+00 0.000000e+00
## SERPINF2 0.0000000 0.000000e+00 2.416595e+00 0.000000e+00 0.000000e+00
## SERTAD3 17.5025175 3.536223e+01 3.159772e+01 3.227628e+01 2.868128e+01
## SETD3 11.2376369 1.337837e+01 7.748257e+00 1.676833e+01 1.050350e+01
## SH3BP1 21.7665502 2.364034e+01 2.209293e+01 3.521958e+01 2.275922e+01
## SHKBP1 38.2388422 4.373535e+01 7.561892e+01 4.613304e+01 6.090089e+01
## SIGLEC10 1.9310320 1.185346e+00 1.864540e+01 3.913710e+00 3.354393e+01
## SIGLEC9 0.0000000 3.458258e-01 2.605346e+01 0.000000e+00 0.000000e+00
## SIK1 10.9254672 7.527678e+00 2.337111e+01 4.635299e+00 1.484658e+01
## SIRPB1 2.4718552 2.170599e+00 1.204132e+01 1.171694e+00 1.438551e+00
## SKAP1 97.3427564 1.036181e+02 9.266438e+00 1.327628e+02 4.005773e+01
## SLC16A3 1.2138911 4.417063e+00 2.337610e+01 2.552814e+00 2.016366e+00
## SLC24A4 0.0000000 0.000000e+00 2.889999e+00 0.000000e+00 0.000000e+00
## SLC35B1 10.3892091 9.895199e+00 1.399944e+01 2.109546e+01 1.145890e+01
## SLC39A3 15.5522745 1.582249e+01 1.281533e+01 2.237620e+01 2.802305e+01
## SLC43A2 0.1486334 1.519294e-01 1.378471e+01 9.158268e-01 2.323529e+00
## SLC44A2 20.4921379 2.006319e+01 1.086482e+01 2.445043e+01 3.101189e+01
## SLC7A7 3.0297858 4.346967e+00 1.276469e+02 4.441923e+00 9.064123e+00
## SLIRP 50.4073060 5.755855e+01 5.798571e+01 4.457885e+01 3.632891e+01
## SMARCB1 8.6821439 6.907556e+00 8.624155e+00 7.811646e+00 2.272323e+01
## SMIM5 0.0000000 0.000000e+00 3.712664e-01 0.000000e+00 0.000000e+00
## SMOX 0.0000000 6.297271e-01 2.440859e+00 0.000000e+00 0.000000e+00
## SMPD3 0.0000000 1.319436e+00 3.335782e-01 6.280047e-01 0.000000e+00
## SNAP23 29.8535025 2.275679e+01 1.971841e+01 2.289211e+01 4.769210e+01
## SNAP29 10.6739633 1.508648e+01 2.616085e+01 1.680804e+01 1.299694e+01
## SNF8 41.1849230 3.874558e+01 7.931308e+01 4.093758e+01 4.239486e+01
## SNN 19.4845583 1.301045e+01 4.023017e+01 2.705604e+00 3.154694e+01
## SNRNP25 31.7693181 1.757247e+01 2.599228e+01 3.474521e+01 3.506647e+01
## SNRPD1 67.0469123 5.139979e+01 4.531791e+01 6.505960e+01 5.814219e+01
## SNRPD2 989.9595052 9.204033e+02 3.637267e+02 9.287777e+02 7.829941e+02
## SNX29P2 5.9938509 2.359804e+00 3.008988e+01 3.443718e+01 4.250794e+02
## SNX5 9.9111392 1.473801e+01 1.172285e+01 2.458915e+01 2.928913e+01
## SOCS3 179.8830293 1.565721e+02 8.187250e+01 6.383139e+01 3.074328e+01
## SOD1 700.2030976 6.842679e+02 2.129016e+02 5.298851e+02 3.718359e+02
## SPECC1 0.4239301 3.978181e-01 9.975490e-01 9.599800e-01 2.836651e-01
## SPG21 44.9938773 4.711497e+01 8.982348e+01 4.234440e+01 4.583654e+01
## SPHK1 0.4463119 0.000000e+00 4.526641e+00 0.000000e+00 0.000000e+00
## SPIB 3.0637056 1.014360e+00 2.673064e+00 7.527741e-01 1.136893e+02
## SPINT1 1.2825151 1.977380e+00 5.503791e+00 1.141484e+00 6.076416e+00
## SPINT2 46.4677108 2.421263e+01 5.002293e+01 1.307852e+01 8.309054e+01
## SPN 14.3483073 2.332285e+01 2.221238e+01 3.603423e+01 5.948801e+00
## SPPL2A 1.4878874 3.445852e+00 3.827000e+00 5.312194e+00 2.818022e+00
## SPTSSA 10.7291308 1.238763e+01 5.053866e+01 3.091707e+01 2.416112e+01
## SRSF5 473.0944599 3.964246e+02 2.704138e+02 3.453318e+02 4.117422e+02
## ST13 245.4963158 1.958990e+02 8.755918e+01 1.612342e+02 1.774855e+02
## STX10 70.6489035 8.048152e+01 1.855273e+02 5.761384e+01 9.499516e+01
## STXBP2 21.6302524 2.968151e+01 1.426922e+02 3.495539e+01 2.820462e+01
## SULF2 2.3078393 2.307532e+00 1.309472e+01 1.794681e+00 5.721690e+00
## SULT1A1 15.1561757 1.615646e+01 1.273348e+02 4.119796e+00 1.774337e+00
## SUMO3 91.2611653 1.095874e+02 1.180272e+02 9.886931e+01 2.000124e+02
## SUN2 47.9313232 4.942825e+01 1.693929e+01 8.250101e+01 3.222120e+01
## SUPT4H1 121.9033892 1.287323e+02 3.426705e+02 2.137312e+02 1.816936e+02
## SYNE2 2.6291375 4.874948e+00 4.050958e-01 9.520382e+00 2.851804e+00
## SYNGR1 1.2994289 3.388603e-01 1.205077e+01 7.373832e+00 0.000000e+00
## SYNGR2 18.4714380 2.603994e+01 5.894793e+01 2.450652e+01 8.167143e+01
## TAX1BP3 30.6600832 4.046328e+01 8.668177e+01 4.905183e+01 4.784985e+01
## TBCB 91.1575627 1.330178e+02 1.099035e+02 2.203310e+02 1.172489e+02
## TBX21 8.5784757 6.234356e+00 5.199089e+00 5.885117e+01 1.059240e+01
## TBXA2R 2.7770801 7.761678e-01 0.000000e+00 0.000000e+00 4.669888e+01
## TCF4 0.2802272 3.375060e-01 8.578613e-01 3.105312e-01 6.768587e+00
## TCL1A 15.1123866 1.242909e+01 9.947854e+00 1.286121e+01 1.597364e+03
## TECR 62.1197637 6.808303e+01 1.966723e+01 7.172540e+01 3.143896e+01
## TGFB1 41.3073108 7.010511e+01 1.384736e+02 1.087023e+02 3.439104e+01
## TGFB1I1 0.0000000 4.200458e-01 2.797886e-01 4.777243e-01 1.508457e+00
## TIMM13 137.5381371 1.238933e+02 2.944220e+02 1.442124e+02 1.544037e+02
## TMC6 16.1021251 1.544744e+01 6.995423e+00 2.037995e+01 1.164823e+01
## TMED1 21.9913520 2.199261e+01 2.672043e+01 3.313962e+01 1.758057e+01
## TMED3 6.8850639 4.521180e+00 5.363474e+00 5.741696e+00 6.168288e+00
## TMEM11 41.9784680 4.325286e+01 3.981642e+01 5.296563e+01 4.550149e+01
## TMEM205 30.6068103 5.043830e+01 2.447732e+02 1.421477e+02 8.899424e+01
## TMEM219 162.1039333 2.023414e+02 3.793292e+02 1.167776e+02 1.141203e+02
## TMEM91 24.3522901 6.819734e+00 3.933512e+01 1.428774e+01 9.421401e-01
## TMIGD2 134.7548024 3.379595e+01 4.992186e+00 8.167297e+01 4.856947e+00
## TMSB4Y 0.0000000 1.085946e+00 9.252727e+00 3.433371e+00 1.376179e+01
## TNFAIP2 2.0574387 2.117392e+00 6.930636e+01 0.000000e+00 1.237553e+00
## TNFSF13B 9.7923639 3.104681e+01 3.155864e+02 1.695275e+01 1.310274e+01
## TNNC2 2.3736855 0.000000e+00 6.329959e+00 0.000000e+00 3.730925e+00
## TOMM40 20.9442634 3.029014e+01 1.356198e+01 2.642127e+01 1.792955e+01
## TOP1 7.1664636 7.499989e+00 6.664586e+00 9.676750e+00 1.078988e+01
## TPM1 1.1503657 2.814482e+00 2.575694e+00 2.801246e+00 8.315479e-01
## TPM4 5.5895202 1.455494e+01 1.431473e+01 1.446386e+01 1.297216e+01
## TPPP3 0.7119928 3.072481e+00 3.572698e+01 1.490378e+00 0.000000e+00
## TPST2 11.8161238 2.219104e+01 1.212025e+01 8.223518e+01 1.089685e+01
## TPTEP1 35.9019896 9.166703e+00 6.957765e+01 1.285716e+01 6.570807e+00
## TRADD 142.2304770 2.660108e+02 4.987988e+01 1.067058e+02 6.543323e+01
## TRAPPC1 524.0162140 6.019193e+02 6.650442e+02 5.875431e+02 6.579235e+02
## TRAPPC5 7.6477433 6.376577e+00 1.855883e+01 5.912725e+00 9.112104e+00
## TSEN34 16.6003347 3.176260e+01 5.430020e+01 2.399976e+01 2.568513e+01
## TSEN54 19.5334352 1.476706e+01 3.067472e+00 4.624535e+01 8.243354e+00
## TSPAN3 5.3404307 4.761617e+00 3.831333e+00 5.932507e+00 3.123865e+01
## TSPO 365.5779114 5.490762e+02 1.689902e+03 3.886786e+02 3.045868e+02
## TTC38 3.3324981 9.667629e-01 2.900001e+00 4.709571e+01 6.175921e+00
## TTC39C 38.5151344 4.957884e+01 4.708060e+00 2.982803e+01 1.157006e+01
## TUBA8 0.0000000 4.011863e-01 0.000000e+00 0.000000e+00 0.000000e+00
## TUBB1 8.7343856 5.104551e+00 5.852358e+00 4.733511e+00 1.404787e+00
## TUBB6 0.0000000 5.720548e-01 9.592386e+00 1.692770e+00 2.732806e+01
## TXN2 131.6279173 1.145071e+02 1.231054e+02 1.261725e+02 1.096252e+02
## TXNDC17 38.8121773 2.962363e+01 6.397896e+01 2.867005e+01 2.716951e+01
## TXNL1 22.6305063 2.121734e+01 1.594967e+01 1.715365e+01 2.400896e+01
## TYMP 43.4868099 7.243612e+01 1.017822e+03 5.567958e+01 7.235026e+01
## TYROBP 197.6528304 1.655470e+02 1.249587e+04 4.874533e+02 1.714280e+02
## UBB 1364.7030864 1.570818e+03 9.094749e+02 1.952545e+03 1.711737e+03
## UBL5 338.0855656 3.820258e+02 4.822721e+02 4.398279e+02 2.538916e+02
## UBL7 50.6499790 5.904733e+01 3.920317e+01 4.768175e+01 7.536128e+01
## UNC119 13.7193525 1.384081e+01 2.067173e+01 2.028708e+01 2.459025e+01
## UPK3A 0.0000000 0.000000e+00 2.649908e+01 0.000000e+00 1.473125e+00
## UQCR10 492.1028947 4.753514e+02 6.226033e+02 4.936918e+02 3.506080e+02
## UQCRFS1 103.4693159 1.006231e+02 1.710663e+02 1.032926e+02 8.228948e+01
## USF2 24.4882276 2.758724e+01 2.291786e+01 2.174391e+01 4.482077e+01
## USP10 29.1242273 5.480467e+01 6.919249e+00 1.009328e+01 1.021017e+01
## UTY 5.0779158 4.223884e+00 4.227210e+00 6.920024e+00 5.137416e+00
## VASP 59.9397570 8.332362e+01 1.437706e+02 9.819513e+01 1.003516e+02
## VMO1 0.0000000 0.000000e+00 1.780337e+00 0.000000e+00 0.000000e+00
## VMP1 11.5132963 1.485237e+01 2.027370e+01 1.226921e+01 1.024764e+01
## VPREB3 14.8114914 1.179059e+01 2.441713e+01 1.017998e+01 2.316307e+03
## VPS35 9.8061613 1.258279e+01 7.809647e+00 8.194302e+00 1.268809e+01
## VSTM1 0.0000000 1.991723e+00 2.146480e+02 8.309776e+00 8.746170e+00
## WDR83OS 524.5958924 5.955096e+02 1.114003e+03 7.532722e+02 7.059644e+02
## WIPI1 0.8968096 3.031535e+00 1.010117e+01 1.170113e+01 3.456312e+00
## WSB1 24.3984250 2.414157e+01 1.992704e+01 2.892851e+01 2.763692e+01
## XBP1 142.2843478 1.801308e+02 7.827202e+01 2.507621e+02 1.029273e+02
## YIF1B 12.4873211 1.901290e+01 2.365932e+01 1.515940e+01 5.549853e+00
## YPEL1 2.7659986 4.752453e+00 1.074865e+00 1.519902e+01 1.677234e+01
## YWHAE 25.3889197 4.805931e+01 5.695952e+01 4.078123e+01 6.482539e+01
## YWHAH 64.9397330 1.257939e+02 1.148723e+02 1.231499e+02 8.547854e+01
## ZDHHC1 0.5775906 5.067445e-01 2.041007e+01 1.750737e+00 0.000000e+00
## ZFAND6 32.9166145 3.968806e+01 1.867157e+01 5.964291e+01 5.777733e+01
## ZFP36 884.8475750 1.370694e+03 1.527962e+03 1.000149e+03 1.030806e+03
## ZFP36L1 49.0704063 7.301232e+01 5.120933e+01 4.618903e+01 1.035364e+02
## ZGLP1 1.4561907 0.000000e+00 2.226570e+01 1.938630e+00 2.267520e+00
## ZMAT5 17.4070130 2.066098e+01 1.871741e+01 3.882692e+01 2.640098e+01
## ZMYND8 8.3259328 4.908424e+00 1.879018e+00 4.650267e+00 4.030965e+00
## ZNF106 6.1899355 5.735979e+00 1.389016e+01 4.387477e+00 6.350533e+00
## ZNF689 3.7297223 3.573070e+00 2.884895e+00 3.054414e+00 2.535071e+00
## ZNF778 0.0000000 0.000000e+00 1.430040e-01 3.060152e-01 3.023665e-01
## ZNHIT3 37.5217212 3.386952e+01 1.978637e+01 4.452879e+01 3.084430e+01
## FCGR3A_Mono NK DC Platelet
## AAK1 1.855434e+00 9.797306e+00 5.872226e-01 0.000000e+00
## AAMP 5.397468e+01 5.838583e+01 8.719621e+01 0.000000e+00
## ABCF3 9.058885e+00 1.263112e+01 2.110337e+01 0.000000e+00
## ABHD14B 2.938013e+01 8.677067e+01 4.063948e+01 0.000000e+00
## ABHD16A 1.736920e+01 8.376243e+00 1.920409e+01 1.709078e+02
## ABHD5 1.244156e+01 2.984585e+00 3.550575e+00 0.000000e+00
## ACAA1 6.767393e+01 2.372511e+01 5.836643e+01 0.000000e+00
## ACTR2 9.123542e+01 7.815007e+01 1.230117e+02 5.380564e+00
## ACTR3 5.685667e+01 9.280679e+01 4.674879e+01 2.980605e+01
## ADAR 3.242310e+01 2.061603e+01 1.444733e+01 1.690210e+00
## ADH5 5.864707e+01 8.251666e+01 2.294892e+01 7.398344e+01
## ADI1 5.377792e+01 4.699932e+01 6.581355e+01 2.040888e+02
## AFF3 6.439730e-01 7.365448e-02 4.665672e+00 0.000000e+00
## AGPAT1 6.586551e+00 1.642388e+01 1.972090e+01 1.525484e+02
## AGTRAP 1.668934e+02 1.053550e+02 2.113919e+01 7.821681e+01
## AIF1 8.333150e+03 1.183497e+02 2.074640e+03 6.247564e+02
## AKR1A1 6.502038e+01 6.324239e+01 1.107820e+02 3.597833e+01
## AMPD2 1.665446e+01 3.985207e+00 1.395515e+01 3.324628e+01
## ANXA4 2.919797e+01 3.429434e+01 1.466930e+01 3.931801e+00
## ANXA5 4.905036e+02 5.237468e+01 3.699176e+02 5.325609e+01
## ANXA6 2.998728e+01 1.965022e+02 4.952262e+01 3.571159e+01
## ARHGAP15 1.610089e+01 6.517425e+01 5.298540e+01 0.000000e+00
## ARHGAP24 4.449412e+00 0.000000e+00 1.898588e+00 0.000000e+00
## ARHGEF3 6.315221e+00 4.677612e+01 0.000000e+00 0.000000e+00
## ARL4C 1.178201e+01 2.272099e+02 1.993347e+02 7.114100e+01
## ARL6IP5 2.637235e+02 4.245914e+02 1.798850e+02 9.935037e+02
## ARPC2 6.601796e+02 7.160938e+02 4.619098e+02 4.572967e+02
## ARPC4 2.035271e+02 3.668546e+02 1.903367e+02 2.035578e+02
## ARPC5 1.632335e+02 1.507961e+02 1.276978e+02 5.367764e+02
## ARSB 1.318231e+00 0.000000e+00 8.523596e+00 0.000000e+00
## ASAP2 0.000000e+00 0.000000e+00 2.025480e+00 1.761654e+01
## ATF3 2.758908e+01 7.396348e+00 4.780819e+01 0.000000e+00
## ATG3 1.064862e+02 2.756473e+01 9.150941e+01 2.093237e+01
## ATOX1 5.649870e+01 5.598621e+01 3.698749e+01 6.091460e+01
## ATP1A1 6.775198e+01 2.562219e+01 1.635207e+01 5.838474e+01
## ATP1B3 2.515308e+02 2.076339e+02 1.149256e+02 8.308314e+00
## ATP6V0B 3.287660e+02 1.635957e+02 2.319446e+02 5.480609e+01
## BAG6 2.480932e+01 3.498588e+01 2.078984e+01 0.000000e+00
## BANK1 4.589002e+00 1.251412e+01 6.954557e+00 1.373007e+01
## BASP1 2.307332e+01 3.351268e+00 1.253899e+02 0.000000e+00
## BATF3 1.099135e+02 0.000000e+00 3.866665e+01 0.000000e+00
## BCL11A 2.515704e-01 7.098444e-01 5.893554e+00 0.000000e+00
## BHLHE40 9.001060e+00 3.835524e+01 5.489381e+01 2.399247e+01
## BIN1 6.702099e+00 6.465840e+01 4.833337e+01 0.000000e+00
## BRK1 1.305223e+03 1.031288e+03 1.240564e+03 2.181229e+03
## BST1 4.439891e+01 2.987152e+00 3.087870e+01 0.000000e+00
## BTG2 1.189202e+02 2.281603e+02 8.011783e+01 7.886420e+00
## BTN3A2 2.754180e+01 8.927235e+01 7.986543e+00 3.101232e+01
## BZW1 1.661139e+01 7.542703e+01 3.376596e+01 5.378567e+00
## C1orf122 4.630360e+01 1.184393e+02 2.456924e+01 9.745791e+00
## C1orf162 6.305451e+02 4.563831e+02 9.530562e+02 7.855079e+00
## C1orf198 0.000000e+00 3.875780e+00 0.000000e+00 7.026106e+01
## C1orf21 3.383389e-01 4.370970e+01 9.671916e-01 0.000000e+00
## C1orf43 1.624452e+02 1.141845e+02 1.855898e+02 3.412016e+02
## C1QA 9.697652e+01 0.000000e+00 5.567768e+00 4.253347e+01
## C2orf88 6.541106e-01 0.000000e+00 0.000000e+00 4.237045e+02
## CACNA2D3 0.000000e+00 1.871604e+00 4.857518e+01 0.000000e+00
## CALM2 3.049264e+02 1.521311e+02 2.627510e+02 9.693336e+01
## CAMK1 1.501309e+02 0.000000e+00 4.056603e+01 1.601684e+01
## CAMK4 4.258597e+00 2.480828e+00 5.283256e+00 1.597096e+00
## CAP1 2.743941e+02 3.821260e+02 2.299451e+02 2.159325e+02
## CAPG 1.656606e+02 3.758645e+01 5.185180e+02 8.988367e+00
## CAPN2 3.292421e+01 4.856316e+01 5.083628e+01 1.150730e+02
## CAPZB 8.019585e+01 8.503326e+01 4.554262e+01 1.285785e+02
## CAST 1.104192e+01 3.417122e+01 1.448667e+01 1.311420e+01
## CCDC12 2.047110e+02 1.732893e+02 5.064905e+01 4.573593e+00
## CCDC167 6.080378e+01 1.967296e+02 8.188686e+01 0.000000e+00
## CCDC28B 3.297223e+00 7.705009e+01 2.942716e+00 5.613958e+00
## CCDC50 4.862974e+00 3.401471e+00 4.942346e+01 0.000000e+00
## CCDC88A 1.738451e+00 0.000000e+00 6.932520e+00 0.000000e+00
## CCND3 7.249047e+01 2.826444e+02 7.376915e+01 3.503822e+02
## CCNL1 3.308407e+01 3.210103e+01 5.441868e+01 2.490876e+00
## CCR2 3.556560e+00 1.790211e+00 4.155145e+01 0.000000e+00
## CCT5 7.767174e+01 7.699583e+01 1.049385e+02 3.856484e+00
## CD14 7.108425e+01 1.468252e+01 7.646406e+01 6.817751e+01
## CD160 0.000000e+00 1.601724e+02 0.000000e+00 0.000000e+00
## CD180 7.607129e+00 3.090998e+00 1.170307e+01 0.000000e+00
## CD1C 1.658341e+00 4.567208e+00 3.415991e+02 0.000000e+00
## CD1D 1.801719e+01 0.000000e+00 7.430202e+01 0.000000e+00
## CD2 3.171308e+01 2.550682e+02 1.080201e+02 0.000000e+00
## CD244 1.983685e+01 1.032152e+02 4.754067e+00 0.000000e+00
## CD247 3.997187e+00 4.205096e+02 5.428588e+00 0.000000e+00
## CD302 5.065055e+01 4.129595e-01 1.438632e+02 2.754191e+01
## CD38 2.260278e+00 5.292734e+01 1.036247e+01 0.000000e+00
## CD47 4.163008e+01 7.888785e+01 3.977047e+01 1.269271e+02
## CD48 6.078448e+02 4.312657e+02 3.347974e+02 4.009132e+01
## CD52 5.051817e+03 2.271041e+03 2.433667e+03 7.469671e+02
## CD53 4.092117e+02 1.164182e+03 3.593955e+02 3.031068e+01
## CD55 5.821491e+01 2.268943e+01 3.471636e+01 1.246765e+01
## CD74 1.870178e+03 6.845204e+02 1.031014e+04 1.134233e+03
## CD83 8.774162e+01 2.263684e+01 8.655102e+01 0.000000e+00
## CD86 8.727262e+01 8.432880e+00 6.229047e+01 0.000000e+00
## CD8A 5.678837e+00 5.027681e+01 3.850239e+00 0.000000e+00
## CD8B 5.454093e+00 8.882036e+00 8.366094e+00 1.187123e+01
## CD96 6.932623e+00 3.750665e+01 1.518749e+01 0.000000e+00
## CDA 8.750889e+02 2.592236e+01 0.000000e+00 0.000000e+00
## CDC23 2.564688e+00 1.418324e+01 1.105809e+01 0.000000e+00
## CDC42SE1 3.865887e+01 1.396402e+02 5.559285e+01 2.535860e+01
## CDKN1A 5.364464e+01 9.387027e+00 8.172328e+01 1.137765e+02
## CDYL 6.042104e+00 1.147194e+01 6.165647e+00 1.282046e+02
## CEP170 4.695894e-01 7.983371e-01 7.327034e+00 0.000000e+00
## CHCHD5 5.922475e+01 5.974449e+01 7.511450e+01 5.232759e+01
## CHST2 3.464460e+01 8.075466e+01 4.861987e+00 5.274784e+01
## CKS1B 7.518472e+01 5.754343e+01 2.688077e+01 1.858364e+01
## CLIC1 1.133428e+03 2.005993e+03 1.328679e+03 1.213974e+03
## CLIC4 1.572548e+00 4.955459e+00 9.803421e+00 1.141430e+02
## CLTB 1.509814e+02 1.841355e+02 2.894553e+02 1.107634e+01
## CMC1 9.460497e+00 4.381608e+01 1.116758e+01 2.064261e+00
## CMPK2 2.534903e+01 2.704836e+01 3.206117e+01 4.952815e+00
## CMTM6 1.043456e+02 1.198182e+02 5.446834e+01 1.647333e+02
## CMTM7 8.699729e+01 3.036535e+01 4.423828e+01 5.838576e+00
## CMTM8 0.000000e+00 2.145677e+01 0.000000e+00 0.000000e+00
## CNIH4 1.059290e+02 1.689213e+01 1.291842e+01 4.271681e+00
## CNPY3 3.891483e+02 2.966847e+02 7.058683e+02 1.818198e+02
## COA5 1.841389e+01 5.858281e+01 7.894101e+01 6.369978e+01
## COL9A2 9.127014e-01 2.043534e+00 1.806683e+01 0.000000e+00
## COQ2 4.705244e+01 1.206425e+01 2.110479e+01 7.838089e+00
## COX17 2.478094e+02 1.222998e+02 1.479313e+02 1.669972e+02
## COX5B 1.029902e+03 8.942656e+02 9.423850e+02 4.081887e+02
## COX6A1P2 1.085510e+03 1.127368e+03 1.103737e+03 5.114200e+02
## CREG1 3.415116e+01 1.245694e+01 1.910371e+01 2.567180e+01
## CRTAP 1.255098e+01 4.637218e+00 3.414796e+01 3.264694e+00
## CSF1R 1.095212e+02 1.974225e+00 4.562895e+01 1.266328e+01
## CSF3R 1.312786e+01 1.655475e+00 2.506254e+01 1.059924e+01
## CSNK2B 1.644916e+02 2.961893e+02 1.843168e+02 0.000000e+00
## CSRP1 1.277282e+01 5.400013e+01 9.994636e+00 0.000000e+00
## CSTA 5.094842e+01 1.873771e+01 5.534705e+01 0.000000e+00
## CTBS 1.884136e+01 6.043244e+00 1.497917e+01 0.000000e+00
## CTDSP1 2.837245e+01 7.653327e+01 2.846090e+01 9.160674e+00
## CTDSPL 6.383810e+00 0.000000e+00 0.000000e+00 4.784713e+01
## CTNNBIP1 2.178732e+01 3.278667e+01 5.777178e+01 4.014112e+01
## CTSS 1.008591e+03 9.199674e+01 2.262100e+02 8.905736e+01
## CX3CR1 7.426155e+01 1.632776e+02 1.661069e+01 2.378869e+01
## CXCR4 1.332379e+02 4.629203e+02 4.454790e+02 8.208987e+01
## CXXC5 1.993475e+00 8.999283e+01 1.488368e+01 2.958850e+01
## CYB5R1 9.577652e+00 4.850820e+00 1.634518e+01 2.876611e+02
## CYSTM1 6.812186e+01 6.490330e+00 8.554635e+01 0.000000e+00
## CYTIP 2.236077e+02 1.466084e+02 1.388371e+02 1.927558e+01
## DAB2 0.000000e+00 1.694216e+01 7.784930e+00 5.459268e+01
## DAPP1 1.673582e+01 1.795176e+00 3.183547e+01 2.672710e+02
## DBI 2.609705e+02 4.691471e+02 4.255291e+02 7.112056e+01
## DDAH2 9.598701e+01 7.305198e+01 1.594804e+02 4.751940e+01
## DDOST 5.344091e+01 2.520365e+02 9.415696e+01 7.990760e+00
## DEF6 2.342406e+01 8.595130e+01 2.584693e+01 0.000000e+00
## DEGS1 3.499652e+01 6.035051e+01 5.571773e+01 0.000000e+00
## DHX29 2.045979e+01 1.903657e+01 3.182847e+01 0.000000e+00
## DNAJB11 8.915241e+00 3.957789e+01 1.586184e+01 0.000000e+00
## DNASE1L3 1.957904e+00 2.821269e+00 2.296342e+02 0.000000e+00
## DOCK2 1.264010e+01 8.513411e+00 7.200689e+00 0.000000e+00
## DOK3 1.816307e+01 1.386468e+00 3.464200e+00 0.000000e+00
## DRAM2 6.966467e+01 4.803543e+01 6.483712e+01 0.000000e+00
## DUSP2 4.974022e+01 3.043615e+02 2.756854e+02 0.000000e+00
## DUSP23 2.019574e+02 2.598957e+02 6.813094e+02 1.020043e+02
## DYNC1LI1 1.923276e+01 1.479112e+01 1.519039e+01 0.000000e+00
## EAF2 8.327171e+01 1.805002e+01 9.949825e+01 0.000000e+00
## EFHD2 2.393890e+02 4.330119e+02 1.687978e+02 1.716090e+01
## EHD3 0.000000e+00 1.409932e+00 4.549901e+00 3.947315e+01
## EIF4A2 9.906727e+01 2.837633e+02 1.309625e+02 8.140768e+01
## EIF4E2 5.151481e+01 1.682361e+01 2.917198e+01 3.787625e+00
## ENO1 2.647254e+02 3.180515e+02 2.959592e+02 1.569157e+02
## ENSA 7.820698e+01 1.966115e+02 1.021414e+02 9.161118e+01
## EREG 1.228010e+00 0.000000e+00 1.429739e+01 0.000000e+00
## F11R 0.000000e+00 5.028464e+00 8.767715e+00 2.168412e+01
## F13A1 1.266985e+00 5.981573e-01 6.641187e+00 6.256014e+02
## FAM76A 1.135985e+00 6.305904e+00 1.608567e+01 0.000000e+00
## FASLG 0.000000e+00 1.798359e+02 0.000000e+00 0.000000e+00
## FAXDC2 0.000000e+00 0.000000e+00 0.000000e+00 2.325107e+02
## FBLN2 0.000000e+00 0.000000e+00 1.838903e+01 0.000000e+00
## FBXO6 4.635243e+01 1.954122e+02 1.454851e+02 0.000000e+00
## FCER1A 3.444546e+00 2.271262e+01 3.471072e+03 0.000000e+00
## FCER1G 1.006524e+04 3.997088e+03 3.107644e+03 1.681846e+03
## FCGR1A 1.597234e+01 0.000000e+00 8.328547e+00 0.000000e+00
## FCGR2A 7.616304e+01 3.221648e+00 2.952237e+01 7.198723e+01
## FCGR2B 1.341259e+00 1.874907e+00 2.040937e+01 0.000000e+00
## FCGR3A 1.102304e+03 7.204018e+02 1.781547e+01 6.588745e+01
## FCRL1 7.860957e+00 4.479017e+00 0.000000e+00 0.000000e+00
## FCRL2 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
## FCRL6 2.117154e+00 1.664090e+02 1.040143e+01 0.000000e+00
## FCRLA 2.412354e+00 2.942733e+00 9.318082e+00 0.000000e+00
## FDPS 3.421921e+01 1.290015e+02 6.419522e+01 7.275179e+01
## FGD2 1.199049e+01 1.580810e+00 1.415282e+01 0.000000e+00
## FGFBP2 2.608201e+01 5.961928e+03 2.457760e+01 0.000000e+00
## FGR 3.509052e+02 2.442460e+02 1.832081e+02 2.571598e+01
## FHIT 1.068125e+00 3.514718e+00 2.024791e+00 0.000000e+00
## FLOT1 1.665000e+01 1.426799e+02 5.437360e+01 6.177039e+01
## FRMD4B 0.000000e+00 9.527887e-01 9.539433e+00 1.941153e+00
## FSTL1 0.000000e+00 0.000000e+00 0.000000e+00 1.148504e+02
## G0S2 5.787206e+02 3.931584e+01 3.616726e+01 0.000000e+00
## GAPT 3.466534e+01 9.686054e-01 6.676855e+01 0.000000e+00
## GATA2 0.000000e+00 0.000000e+00 0.000000e+00 1.124724e+02
## GBP2 6.226783e+01 7.207264e+01 2.284318e+01 3.010225e+00
## GCA 9.850719e+01 8.412239e+00 4.475617e+01 1.584774e+01
## GCLM 5.974250e-01 3.779715e+01 5.624165e+00 4.007162e+00
## GLRX 1.622529e+02 1.465917e+02 8.453505e+01 1.355130e+01
## GLUL 2.407500e+01 1.113298e+01 9.792208e+00 9.280901e+01
## GMPR 1.344779e+01 0.000000e+00 1.672433e+01 6.231369e+02
## GNAI2 7.330213e+01 5.122829e+01 6.227489e+01 4.842781e+01
## GNAI3 3.202887e+01 2.593393e+01 1.632856e+01 4.556144e+01
## GNB1 2.549748e+01 1.569183e+01 3.710499e+01 3.987342e+01
## GNB4 4.966367e+00 1.423414e+00 9.441098e+00 0.000000e+00
## GNG5 1.694438e+02 1.011311e+02 2.064675e+02 4.346245e+01
## GNL2 9.523191e+00 7.063120e+00 2.638344e+01 0.000000e+00
## GNLY 2.790042e+01 9.020788e+03 5.510314e+01 3.685210e+01
## GP9 3.289539e+00 0.000000e+00 0.000000e+00 9.551731e+03
## GPBAR1 1.468362e+02 1.215544e+01 5.663831e+00 0.000000e+00
## GPR137B 3.981351e+01 3.709662e+00 0.000000e+00 9.424733e+00
## GPX1 7.242125e+02 1.574196e+02 2.654768e+03 2.504323e+04
## GSTM4 0.000000e+00 4.179492e+00 1.261796e+01 5.150334e+01
## GTPBP2 1.297252e+00 4.596215e+00 0.000000e+00 1.269484e+02
## GYPC 3.697576e+01 2.043738e+02 1.065858e+02 3.111343e+01
## GZMA 4.236316e+01 6.954861e+03 0.000000e+00 0.000000e+00
## GZMK 1.840499e+01 2.309945e+02 2.822509e+01 3.999247e+01
## HAVCR2 1.101930e+01 1.606595e+02 3.882637e+01 3.433757e+01
## HCLS1 1.679825e+02 1.461503e+02 1.313778e+02 5.432537e+01
## HDLBP 9.534800e+00 4.469416e+00 4.641700e+00 1.788064e+00
## HENMT1 6.435542e+00 7.866318e+01 2.071819e+01 0.000000e+00
## HES1 5.591732e+01 5.706249e-01 2.697186e+01 0.000000e+00
## HES4 6.071539e+02 1.096468e+01 2.537617e+01 3.889599e+01
## HGD 0.000000e+00 0.000000e+00 0.000000e+00 5.763122e+02
## HIGD2A 2.165065e+03 1.318731e+03 1.565560e+03 2.952987e+02
## HINT1 7.608820e+01 1.655205e+02 1.990353e+02 4.035472e+01
## HK3 3.385937e+01 3.913268e+00 1.092306e+01 6.580768e+00
## HLA-DMA 1.689351e+02 3.322010e+01 1.001961e+03 6.699291e+01
## HLA-DMB 4.505327e+01 1.294420e+01 2.660726e+02 0.000000e+00
## HLA-DOA 1.748529e+00 0.000000e+00 3.878190e+01 0.000000e+00
## HLA-DOB 5.196228e+00 1.342582e+01 6.171583e+01 0.000000e+00
## HLA-DPA1 8.851458e+02 1.074263e+02 2.949694e+03 2.647878e+01
## HLA-DPB1 5.080502e+02 1.039574e+02 3.315636e+03 9.623682e+01
## HLA-DQA1 6.787107e+01 6.576277e+00 9.035426e+02 1.226370e+01
## HLA-DQA2 9.387485e+01 1.279836e+01 1.353765e+03 1.491280e+01
## HLA-DQB1 4.893915e+01 1.359684e+01 1.040157e+03 1.280511e+01
## HLA-DQB2 1.113645e+01 1.222493e+01 1.550160e+02 0.000000e+00
## HLA-DRA 5.458973e+03 6.655701e+02 2.938563e+04 1.529384e+03
## HLA-DRB1 1.349136e+03 1.305724e+02 5.291857e+03 2.864301e+02
## HLA-DRB5 1.467285e+03 1.131828e+02 5.525677e+03 3.216749e+02
## HLX 0.000000e+00 0.000000e+00 1.143814e+01 0.000000e+00
## HMGB2 1.435249e+02 4.356947e+02 1.718021e+02 2.628488e+02
## HNMT 2.486356e+01 0.000000e+00 0.000000e+00 0.000000e+00
## HNRNPA3 4.511988e+01 1.497630e+02 8.088804e+01 2.381502e+01
## HNRNPD 1.275981e+01 2.845960e+01 3.774904e+01 0.000000e+00
## HOPX 9.300425e-01 3.038165e+02 3.694479e+00 0.000000e+00
## HRH2 1.088929e+01 0.000000e+00 3.239033e+01 0.000000e+00
## HSD17B11 1.646726e+02 1.677401e+02 1.161229e+02 9.323696e+00
## HSPE1 1.074156e+02 1.492258e+02 2.005441e+02 0.000000e+00
## ID2 4.254338e+02 1.203952e+03 7.197784e+02 6.056539e+01
## IFI44 5.996742e+01 9.777233e+01 1.276545e+02 0.000000e+00
## IFI6 1.412341e+03 7.021252e+02 1.756316e+03 1.199662e+02
## IFT57 5.954539e+01 2.030290e+01 1.951280e+01 0.000000e+00
## IGFBP7 5.841557e+01 1.158682e+03 1.366739e+02 0.000000e+00
## IL1B 4.829385e+01 4.775521e+00 2.635943e+02 1.267464e+02
## IL6R 6.452414e+00 3.360265e-01 2.484095e+01 8.961563e+00
## IL7R 1.684931e+01 3.937972e+01 3.537418e+01 1.762465e+01
## IQGAP2 1.028139e+01 2.770603e+01 4.679952e+00 2.139835e+00
## IRF1 3.375624e+01 7.679487e+01 1.788954e+01 1.772757e+00
## ISG15 2.787646e+03 1.710523e+03 1.958112e+03 6.606089e+02
## ITM2C 4.244742e+00 1.392840e+01 2.483347e+02 0.000000e+00
## JAK1 2.346110e+01 4.951435e+01 1.291233e+01 5.220157e+01
## JUN 2.176793e+02 7.182277e+02 5.060846e+02 9.796740e+01
## KDM1B 1.885776e+01 3.796522e+00 1.411625e+01 0.000000e+00
## KHDRBS1 6.822862e+01 7.848779e+01 8.532480e+01 3.168407e+01
## KLF3 3.322857e+01 2.311471e+01 1.036694e+01 6.422348e+00
## KYNU 8.862835e+00 1.349592e+00 6.534308e-01 0.000000e+00
## LAMTOR2 3.314968e+02 2.507388e+02 4.224574e+02 7.560821e+01
## LAMTOR5 2.269622e+02 2.181625e+02 2.071924e+02 3.534802e+01
## LAP3 7.461700e+01 4.859903e+01 1.836460e+02 3.546738e+01
## LAPTM4A 3.202892e+02 3.969535e+02 2.146089e+02 4.219183e+01
## LARP1 9.166274e+00 7.699999e+00 8.877220e+00 0.000000e+00
## LCK 1.258525e+01 2.628403e+02 1.618400e+01 0.000000e+00
## LDLRAP1 3.936196e+00 7.512198e+00 5.671824e+00 1.218035e+02
## LEF1 9.533677e+00 1.099765e+01 0.000000e+00 1.168965e+01
## LEPROT 4.147983e+01 1.897527e+01 1.458607e+01 2.555695e+02
## LGALSL 0.000000e+00 0.000000e+00 0.000000e+00 1.877392e+02
## LIMS1 2.678322e+01 1.253916e+01 1.555646e+01 4.510118e+02
## LINC00877 1.841062e+01 1.024585e-01 6.718107e+00 3.852726e+00
## LINC01011 3.405369e+00 8.644323e+00 0.000000e+00 1.034630e+02
## LMNA 5.247131e+00 3.416631e+00 4.607362e+01 9.835443e+01
## LPCAT1 1.015232e+01 5.058581e+01 0.000000e+00 2.538854e+01
## LRPAP1 2.685167e+01 2.844876e+01 1.854755e+01 8.983914e+00
## LRRFIP1 6.475498e+01 4.183791e+01 5.070320e+01 3.113952e+01
## LSM10 2.503961e+02 2.180054e+02 2.202556e+02 1.491213e+02
## LSM6 1.636891e+02 8.050535e+01 5.181690e+01 0.000000e+00
## LST1 5.397024e+03 8.901766e+01 1.196650e+03 2.303387e+02
## LTB 5.740446e+02 3.388901e+02 5.290304e+02 2.177982e+02
## LY6G6F 0.000000e+00 0.000000e+00 0.000000e+00 1.790804e+03
## LY86 2.430719e+02 5.347844e+00 8.208580e+02 1.635401e+02
## LYAR 3.010417e+01 5.214860e+02 5.292632e+01 1.167933e+02
## LYST 1.964505e+01 3.227340e+00 4.362036e+00 1.061913e+01
## MAD2L2 1.870742e+01 6.229767e+01 2.253115e+01 0.000000e+00
## MAGEF1 0.000000e+00 5.806545e+01 1.402732e+02 1.278240e+01
## MAL 8.032644e+00 2.195778e+01 4.036137e+01 0.000000e+00
## MANF 4.111231e+01 2.420352e+02 6.151999e+01 5.859580e+01
## MAPKAPK3 1.253692e+02 3.166040e+01 2.790696e+01 1.110746e+01
## MARCKSL1 3.308597e+02 2.198867e+02 2.089481e+02 1.203969e+02
## MARCO 3.153850e+01 0.000000e+00 6.724000e+00 0.000000e+00
## MBD4 2.068854e+01 4.119825e+01 2.034150e+01 1.869019e+01
## MBNL1-AS1 4.607895e-01 1.113775e+00 3.856340e+00 8.451138e+01
## MCL1 1.035561e+02 9.027971e+01 1.645790e+02 1.048354e+01
## MCUR1 8.340727e+00 8.680016e+00 1.051534e+01 2.641705e+02
## MEF2C 7.558083e+00 3.285231e+00 6.299696e+00 7.176311e+00
## MFSD1 1.435681e+01 1.032130e+01 4.416025e+01 4.812147e+02
## MGAT4A 4.143001e+00 1.340454e+01 1.099033e+01 0.000000e+00
## MGLL 7.612969e+00 4.942451e-01 2.498715e+00 4.135509e+01
## MGST2 7.493724e+00 2.539676e+01 7.147963e+01 0.000000e+00
## MIB2 3.829407e+00 3.553583e+01 0.000000e+00 0.000000e+00
## MNDA 2.108085e+02 5.785788e+00 3.010713e+02 1.436220e+02
## MOB1A 6.971605e+01 4.513769e+01 6.354892e+01 1.258456e+02
## MOB3C 1.836070e+00 6.733926e+00 4.864636e+00 5.907779e+01
## MPZL1 3.387467e+00 1.921808e+00 1.096271e+01 0.000000e+00
## MRPL53 2.647799e+02 1.056952e+02 1.674502e+02 0.000000e+00
## MTHFD2 1.734434e+01 8.869818e+00 1.885611e+01 1.497238e+01
## MTMR11 3.755651e+01 1.669677e+00 3.330545e+01 0.000000e+00
## MTMR14 3.109001e+01 2.153068e+01 3.273136e+01 3.612371e+00
## MXD3 1.923160e+01 8.198713e+00 0.000000e+00 0.000000e+00
## MXD4 6.568416e+00 1.794681e+01 1.757031e+01 0.000000e+00
## MYCL 1.546587e+00 0.000000e+00 4.266734e+01 0.000000e+00
## MYD88 8.488340e+01 5.101531e+01 4.331680e+01 5.390276e+01
## MYLK 9.159264e-02 0.000000e+00 0.000000e+00 7.418069e+00
## MZB1 1.221418e+00 3.607390e+01 1.674773e+02 1.644835e+02
## MZT2A 1.419439e+01 6.503150e+01 1.242859e+02 3.081033e+01
## NAAA 1.630724e+02 2.833091e+01 4.074134e+01 9.847309e+00
## NADK 3.157184e+01 1.133417e+01 7.508392e+00 0.000000e+00
## NAGK 3.410674e+01 1.300101e+01 3.928932e+01 2.443842e+00
## NBPF10 9.417873e+00 6.586035e+00 9.211719e+00 7.391305e+00
## NCF2 1.614137e+02 9.306180e+00 1.238061e+02 0.000000e+00
## NCR3 1.364435e+01 6.609661e+02 0.000000e+00 0.000000e+00
## NDFIP1 5.959904e+01 8.644453e+01 5.867288e+01 1.744809e+02
## NDUFA2 2.602811e+02 3.167402e+02 2.723867e+02 2.720368e+02
## NDUFB3 1.364983e+02 1.325662e+02 1.621892e+02 1.174020e+01
## NDUFB5 9.601734e+01 3.448168e+01 7.653102e+01 2.282937e+01
## NDUFS6 4.359942e+02 4.400648e+02 6.230906e+02 7.353842e+01
## NEXN 5.177390e+00 0.000000e+00 3.108865e+00 1.568248e+02
## NFKBIZ 7.925043e+01 1.720576e+01 2.665366e+01 3.333424e+01
## NMI 8.992286e+01 6.223959e+01 7.015402e+01 6.564873e+00
## NOTCH2 7.926507e+00 2.009551e+00 7.275417e+00 0.000000e+00
## NPL 2.327007e+01 1.658738e+00 0.000000e+00 0.000000e+00
## NPM1 1.323821e+02 2.860053e+02 2.927523e+02 2.628351e+01
## NQO2 4.620033e+01 5.036364e+01 4.877724e+01 0.000000e+00
## NREP 6.489780e-01 1.064767e+00 6.431433e+00 0.000000e+00
## NUDT16 3.503886e+01 1.431759e+01 1.935171e+01 3.093308e+00
## NUDT3 1.798012e+01 6.405502e+00 5.880544e+00 6.330122e+00
## NUP210 3.502107e+00 1.295498e+01 1.453946e+01 0.000000e+00
## OCIAD2 3.785425e+00 1.160410e+02 4.052559e+01 0.000000e+00
## ODC1 2.946161e+01 4.974708e+01 3.423416e+01 1.460407e+03
## ODF2L 6.337196e+00 2.628964e+01 1.202872e+00 0.000000e+00
## OST4 1.334426e+03 1.573732e+03 1.337317e+03 9.412748e+03
## OXNAD1 5.366540e+00 1.171216e+01 4.525324e+00 0.000000e+00
## P2RY13 2.735744e+01 4.388569e+00 7.991781e+01 0.000000e+00
## PAFAH2 0.000000e+00 8.547240e+00 1.988399e+01 0.000000e+00
## PAIP1 3.377305e+01 2.819706e+01 5.003593e+01 0.000000e+00
## PAPSS1 2.959160e+01 2.472066e+01 4.886653e+00 0.000000e+00
## PARL 6.313201e+01 7.124222e+01 9.895525e+01 1.917360e+01
## PARP1 4.252222e+00 1.189266e+01 6.635900e+00 0.000000e+00
## PBX1 0.000000e+00 0.000000e+00 0.000000e+00 4.540608e+00
## PBXIP1 1.907321e+01 6.508357e+01 0.000000e+00 0.000000e+00
## PDCL3 1.315485e+02 8.569177e+01 1.160343e+02 4.090614e+01
## PDIA6 8.324133e+01 1.576263e+02 1.181014e+02 2.591649e+01
## PDLIM5 1.137712e+01 4.423434e+00 6.820918e+00 0.000000e+00
## PDZK1IP1 3.053560e+00 0.000000e+00 0.000000e+00 9.472056e+02
## PF4 3.781706e+01 0.000000e+00 5.869727e+01 4.102659e+04
## PGD 5.208103e+01 1.395836e+01 5.840544e+01 2.778631e+02
## PHACTR1 2.520754e+00 0.000000e+00 1.555583e+01 8.878260e-01
## PID1 2.855169e+00 0.000000e+00 4.063684e+01 0.000000e+00
## PIM1 2.008815e+01 2.022762e+02 1.964185e+01 7.175861e+00
## PLA2G12A 5.256571e+00 1.031823e+01 1.410390e+01 3.804819e+02
## PLAC8 2.462295e+02 9.768690e+02 4.085800e+02 3.539245e+01
## PLEK 1.199118e+02 3.178069e+02 1.141380e+02 3.993002e+02
## PLEKHO1 2.784552e+01 9.115198e+00 3.941560e+01 7.768096e+01
## PLSCR1 7.566251e+01 7.321456e+01 1.174394e+02 0.000000e+00
## PMVK 3.853523e+02 1.472292e+02 2.230792e+02 1.337637e+02
## POLE4 2.871323e+02 8.514076e+01 7.757594e+01 1.645320e+02
## PPBP 5.164451e+01 1.244306e+00 1.395677e+01 8.335303e+04
## PPCS 9.609631e+01 1.147931e+02 3.014726e+01 3.111194e+01
## PPP1R18 6.608043e+01 2.817057e+02 3.515565e+01 4.727736e+00
## PPP3R1 1.346881e+01 2.622800e+01 4.958859e+01 6.463394e+00
## PPT1 1.453663e+02 4.301584e+01 2.311206e+02 5.519977e+01
## PRADC1 8.893299e+01 7.340490e+01 5.076700e+01 1.548637e+01
## PRDX1 5.302187e+02 3.814794e+02 2.958074e+02 1.417606e+02
## PRDX6 1.737974e+02 1.735527e+02 2.294387e+02 2.162714e+03
## PRELID1 1.221809e+03 5.556921e+02 9.721053e+02 9.175116e+01
## PRKCD 2.652456e+01 1.401962e+01 2.815890e+01 7.461490e+01
## PRPF40A 1.462391e+01 4.232665e+01 2.785000e+01 0.000000e+00
## PSMA5 6.417694e+01 1.212080e+02 7.982853e+01 1.471181e+01
## PSMB4 1.271533e+02 8.301843e+01 1.180910e+02 1.023193e+01
## PSMB8 2.629576e+02 5.385391e+02 2.776655e+02 6.919114e+01
## PSMB9 5.203565e+02 5.684148e+02 3.158567e+02 1.602588e+02
## PSRC1 1.384172e+00 2.064537e+00 0.000000e+00 6.519567e+01
## PTCRA 8.942621e+00 0.000000e+00 8.352792e+01 6.309547e+03
## PTP4A2 9.425038e+01 6.511675e+01 6.309682e+01 1.370016e+02
## PTPN18 5.263385e+01 4.540775e+01 6.197679e+01 2.752302e+02
## PTPN4 1.848618e+00 2.958038e+01 1.025193e+00 1.637757e+00
## PTPN7 4.592102e+00 1.840189e+02 1.018802e+01 0.000000e+00
## PYHIN1 9.705799e+00 1.050128e+02 0.000000e+00 0.000000e+00
## QPCT 7.971537e+00 4.709586e+00 3.363433e+01 0.000000e+00
## RAB10 1.056445e+02 3.725195e+01 4.830589e+01 3.926338e+02
## RAB13 1.004181e+00 0.000000e+00 3.834919e+01 6.381556e+01
## RAB24 1.443473e+02 7.575124e+01 3.953620e+01 0.000000e+00
## RABGAP1L 1.688552e+01 8.825536e+00 4.757241e+00 7.914594e+01
## RAD50 4.704424e+00 1.045842e+01 1.428317e+01 0.000000e+00
## RALB 7.573660e+01 2.306034e+01 2.432663e+01 1.155206e+02
## RALGPS2 2.632346e+00 2.285845e+00 3.238466e+00 0.000000e+00
## RAMP1 0.000000e+00 3.608299e+02 0.000000e+00 0.000000e+00
## RASSF5 3.721570e+01 7.917254e+01 1.973204e+01 0.000000e+00
## RBP7 2.120854e+02 1.341588e+00 0.000000e+00 0.000000e+00
## RCAN3 4.812852e+00 4.825745e+00 3.179181e+01 0.000000e+00
## RCC2 7.581308e+00 2.869320e+01 2.968003e+01 0.000000e+00
## RCSD1 2.786676e+01 4.212083e+01 3.502808e+01 1.065174e+01
## REEP5 1.962559e+02 1.025887e+02 8.340332e+01 4.730866e+01
## REL 8.219310e+00 1.239983e+01 2.554200e+01 4.924837e+00
## RER1 7.096799e+01 8.897163e+01 5.243734e+01 0.000000e+00
## RETSAT 3.258468e+00 8.670692e+00 1.760879e+01 0.000000e+00
## RGS1 1.294838e+01 3.405752e+01 1.320118e+02 0.000000e+00
## RGS18 2.408100e+01 2.688123e+00 2.828934e+01 1.937251e+03
## RGS2 7.115444e+02 1.725640e+02 5.908501e+02 0.000000e+00
## RHOA 3.690953e+02 3.306188e+02 2.448868e+02 4.454982e+02
## RHOB 2.297559e+02 1.599287e+01 7.557485e+01 1.837166e+01
## RHOC 4.849845e+02 3.029658e+02 7.199172e+01 2.313139e+02
## RHOH 7.263376e+00 3.769499e+01 1.131194e+01 0.000000e+00
## RNF11 1.458052e+01 3.432478e+01 2.362351e+01 6.500250e+02
## RNF13 9.346121e+01 4.121973e+01 6.183108e+01 0.000000e+00
## RNF130 4.364362e+01 7.583115e+00 6.203143e+01 5.089223e+00
## RNF144B 2.606577e+01 1.456088e+00 0.000000e+00 3.012455e+01
## RNF149 7.632774e+01 3.614934e+01 1.974346e+01 1.224720e+01
## RNF181 3.645660e+02 2.409452e+02 3.270358e+02 0.000000e+00
## RNF7 1.221708e+02 1.256172e+02 1.504478e+02 0.000000e+00
## RNPEP 7.756200e+01 4.488615e+01 4.933738e+01 0.000000e+00
## RPA2 4.603028e+01 2.389939e+02 4.651979e+01 0.000000e+00
## RPL22 2.533331e+02 2.874343e+02 5.642384e+02 1.809455e+01
## RPL22L1 9.846852e+01 1.303323e+02 2.323076e+02 4.115605e+01
## RRAGC 1.599947e+01 1.131699e+01 2.952038e+01 4.249972e+00
## RSAD2 2.703144e+01 1.717702e+01 2.013985e+00 0.000000e+00
## RSPH9 2.687614e+00 0.000000e+00 0.000000e+00 1.630532e+02
## RTN4 1.196818e+02 6.485470e+01 5.872540e+01 7.823555e+01
## RUFY1 9.600598e+00 1.229623e+01 5.398622e+00 9.067852e+02
## RUNX3 2.151480e+01 7.427457e+01 1.783475e+01 2.723105e+01
## S100A10 2.594513e+03 1.444114e+03 4.035548e+03 2.562921e+02
## S100A11 9.664970e+03 1.800963e+03 3.964936e+03 6.755489e+02
## S100A12 3.290572e+01 6.176737e+01 6.628662e+01 0.000000e+00
## S100A4 1.914911e+04 7.110382e+03 8.413598e+03 2.315330e+03
## S100A6 9.382840e+03 2.663064e+03 5.220504e+03 1.731547e+03
## S100A8 1.210140e+03 1.397254e+02 6.650973e+02 6.136867e+02
## S100A9 5.348253e+03 3.411903e+02 4.842022e+03 1.879094e+03
## SCARB2 4.012180e+00 7.885128e-01 2.336135e+00 2.013177e+01
## SCFD2 6.785433e+00 4.796345e+00 4.777152e+00 2.490209e+02
## SCP2 4.265174e+01 1.689700e+02 6.334074e+01 9.154749e+01
## SCYL3 4.579746e-01 6.170252e+00 1.095263e+01 0.000000e+00
## SDF4 1.948244e+01 1.177457e+02 3.504007e+01 4.639961e+00
## SDHB 1.174465e+02 1.132092e+02 1.353762e+02 1.263218e+02
## SEC24D 3.046028e+00 1.639423e-01 8.290206e+00 0.000000e+00
## SEC62 2.729413e+01 3.924401e+01 1.652214e+01 4.776541e+01
## SEL1L3 8.340142e-01 5.104618e+00 7.702933e+00 0.000000e+00
## SELL 9.500213e+01 5.719284e+02 2.750090e+02 7.996659e+01
## SEMA4A 1.322928e+01 6.014103e-01 2.453424e+01 0.000000e+00
## SERP1 2.939679e+02 1.588365e+02 2.443256e+02 2.034669e+02
## SERPINB1 2.300950e+02 3.552616e+02 2.003766e+02 3.329745e+02
## SERPINB6 2.800319e+01 2.528502e+01 3.050995e+01 2.842554e+01
## SERPINE2 0.000000e+00 1.267358e+00 0.000000e+00 6.449197e+01
## SF3B4 7.243711e+01 1.943357e+02 1.422568e+02 4.953701e+01
## SH2D1B 1.154118e+01 1.789422e+02 0.000000e+00 0.000000e+00
## SH2D2A 0.000000e+00 1.626052e+02 5.498199e+00 0.000000e+00
## SH3BGRL3 6.338734e+03 6.889873e+03 6.540274e+03 2.508978e+04
## SH3BP2 1.192324e+01 1.117820e+01 9.564172e+00 7.943994e+00
## SH3BP5 9.040556e+00 8.804374e+01 1.407597e+01 0.000000e+00
## SH3D19 3.746528e+00 4.923173e+00 1.153964e+01 0.000000e+00
## SH3GLB1 4.779452e+01 5.877753e+01 1.905797e+01 1.541933e+01
## SH3RF1 3.501311e+00 0.000000e+00 1.416239e+01 0.000000e+00
## SH3YL1 1.632182e+00 9.588073e+00 7.949623e+00 1.320749e+01
## SKP1 5.940612e+01 8.400857e+01 5.783384e+01 1.405345e+02
## SLAMF7 1.075134e+01 5.459046e+01 1.606163e+01 0.000000e+00
## SLC11A1 4.753542e+01 1.872996e+00 3.767851e+00 0.000000e+00
## SLC30A9 2.264071e+00 6.984204e+00 9.989410e+00 0.000000e+00
## SLC39A1 4.234371e+01 2.436948e+01 3.383638e+01 0.000000e+00
## SLC40A1 7.462712e+00 5.147839e+00 1.068927e+01 4.570899e+02
## SMAP2 7.632440e+01 3.782489e+01 6.375173e+01 6.038006e+00
## SMIM14 2.108303e+01 1.376077e+01 2.506149e+01 0.000000e+00
## SNAPIN 1.128836e+02 2.450826e+02 5.787265e+01 1.296893e+02
## SNCA 4.731436e+00 4.351852e+00 1.103833e+01 4.171739e+02
## SNX18 2.193225e+01 2.872601e+00 1.107289e+01 3.910192e+01
## SNX2 5.290455e+01 1.805879e+01 1.945345e+01 4.098412e+01
## SNX27 6.963033e+00 1.246251e+00 2.418078e+00 0.000000e+00
## SP110 5.090075e+01 3.203243e+01 3.907796e+01 2.034896e+00
## SPARC 4.082604e+00 0.000000e+00 0.000000e+00 1.837129e+03
## SPATS2L 2.366671e+00 1.125602e+00 1.914882e+01 0.000000e+00
## SPOCD1 0.000000e+00 0.000000e+00 0.000000e+00 1.530331e+02
## SPON2 2.514976e+00 4.991068e+02 5.760252e+00 0.000000e+00
## SRA1 7.437187e+01 5.968018e+01 8.402693e+01 0.000000e+00
## SRD5A3 1.631170e+01 1.238094e+01 1.619142e+01 0.000000e+00
## SSR3 7.152300e+01 3.932076e+01 7.211337e+01 1.210045e+01
## SSX2IP 4.129604e-01 1.372085e-01 3.229707e+00 3.463253e+01
## ST3GAL3 1.929289e-01 0.000000e+00 0.000000e+00 3.979477e+01
## ST3GAL5 7.782699e+00 6.076645e+00 1.818454e+00 1.333918e+00
## ST3GAL6 6.433823e-01 0.000000e+00 1.504388e+01 0.000000e+00
## STAT4 5.582509e+00 3.256045e+01 4.994170e+00 0.000000e+00
## STMN1 2.116458e+00 1.039802e+02 2.718097e+01 1.003173e+01
## STX18 9.416062e+00 1.462511e+01 2.741351e+01 0.000000e+00
## SYTL1 5.801381e+01 9.846848e+01 2.244742e+01 7.787557e+00
## TADA3 4.484877e+01 8.440712e+01 5.091725e+01 1.122206e+02
## TAGLN2 3.870319e+02 5.513440e+02 1.010902e+03 5.965638e+03
## TANK 3.315159e+01 2.752039e+01 1.580650e+01 0.000000e+00
## TAP1 2.680465e+01 1.398137e+02 7.449432e+01 1.781576e+01
## TARBP1 2.322045e+00 4.175134e+00 1.675530e+01 0.000000e+00
## TBC1D9 0.000000e+00 2.779538e-01 1.503849e+01 0.000000e+00
## TCF7 1.126409e+01 1.092654e+01 1.213012e+01 6.209076e+00
## TFDP2 2.312996e+00 1.800435e+01 1.271685e+00 0.000000e+00
## TGFBI 3.786944e+01 1.872431e+00 5.408775e+01 5.635055e+00
## THEMIS2 9.121550e+01 6.812628e+01 5.758561e+01 9.410456e+00
## TIGIT 1.101476e+00 8.138002e+01 2.353761e+00 0.000000e+00
## TKT 1.979675e+02 4.315162e+01 6.942411e+01 2.554678e+01
## TLR10 0.000000e+00 0.000000e+00 1.620131e+01 0.000000e+00
## TMCC2 0.000000e+00 0.000000e+00 2.736257e+00 3.316246e+01
## TMED9 5.695621e+01 2.222981e+02 1.576570e+02 1.783894e+01
## TMEM154 1.013612e+01 1.158818e+00 1.174158e+01 0.000000e+00
## TMEM40 1.198608e+00 0.000000e+00 0.000000e+00 1.283063e+03
## TMEM50A 1.046768e+02 2.242071e+02 1.241093e+02 7.200933e+02
## TMEM69 3.676270e+01 3.698727e+01 8.321747e+01 0.000000e+00
## TNFAIP8 1.252827e+01 4.183408e+01 7.378255e+01 1.022472e+01
## TNFRSF14 6.140347e+01 5.148772e+01 3.551915e+01 2.057342e+01
## TNFRSF1B 1.523909e+02 6.215989e+01 3.344322e+01 5.755124e+00
## TNFRSF25 6.059594e+00 2.325375e+01 0.000000e+00 0.000000e+00
## TNFRSF4 0.000000e+00 3.883132e+01 7.660375e+00 0.000000e+00
## TNFRSF8 2.661435e+01 0.000000e+00 0.000000e+00 4.784020e+01
## TNFSF10 3.315841e+02 5.873467e+01 6.366676e+01 2.527505e+01
## TNFSF4 3.002744e-01 0.000000e+00 0.000000e+00 1.811514e+01
## TNS1 0.000000e+00 0.000000e+00 0.000000e+00 2.783134e+01
## TPM3 9.665363e+01 8.082339e+01 8.531843e+01 2.811928e+02
## TPMT 5.464886e+01 4.160461e+01 5.041590e+01 0.000000e+00
## TRABD2A 2.881307e+00 8.367106e-01 3.449832e+00 0.000000e+00
## TRAF3IP3 5.775914e+01 1.258750e+02 6.458185e+01 1.617244e+01
## TRAK1 2.403321e+00 0.000000e+00 6.694106e+00 0.000000e+00
## TRAPPC12 1.237604e+01 5.330152e+00 8.850511e+00 2.365154e+00
## TRAPPC13 4.717623e+00 9.459408e+00 1.281222e+01 0.000000e+00
## TRAT1 9.687211e+00 2.401093e+01 1.455383e+01 1.170524e+02
## TREM1 5.983839e+00 1.562622e+00 1.586462e+01 0.000000e+00
## TREML1 8.340276e+00 3.341474e+00 0.000000e+00 3.928551e+03
## TREX1 1.724842e+02 6.781046e+01 6.798334e+01 1.501579e+01
## TRIM33 4.770130e+00 1.634661e+00 1.616761e+01 0.000000e+00
## TRIM58 0.000000e+00 0.000000e+00 0.000000e+00 6.000196e+01
## TSTD1 1.053555e+02 3.292581e+02 7.301542e+01 9.380815e+01
## TUBA4A 5.968715e+01 9.858841e+01 1.748928e+01 2.373441e+03
## TUBB2A 4.507266e+00 3.822116e+00 5.056384e+00 2.608426e+02
## TXK 4.370566e+00 1.115727e+02 5.810256e+00 0.000000e+00
## TXNIP 3.723580e+02 1.049993e+03 2.979927e+02 1.074501e+02
## UBE2B 1.287459e+02 7.196048e+01 5.162760e+01 2.314079e+02
## UBE2E2 2.590826e+01 8.813446e+00 5.115877e+01 6.712831e+00
## UBE2F 7.351879e+01 1.439695e+02 4.782238e+01 2.221110e+02
## UBXN11 3.046021e+01 4.361005e+00 1.479257e+01 1.327076e+02
## UNC50 4.949596e+01 1.181462e+02 1.232785e+01 0.000000e+00
## UQCRC1 5.792516e+01 3.697763e+01 4.002939e+01 1.200044e+01
## USP33 7.270284e+00 1.612292e+01 2.694393e+01 0.000000e+00
## VAMP3 5.470427e+01 3.946404e+01 5.512902e+01 0.000000e+00
## VAMP5 7.251555e+02 4.684698e+02 2.672519e+02 5.504521e+01
## VAMP8 9.678892e+02 1.317490e+03 1.850250e+03 3.219426e+02
## VCAN 5.688926e+00 2.430080e+00 1.004841e+01 0.000000e+00
## VDAC1 1.367446e+02 1.074016e+02 8.705394e+01 2.055369e+01
## VIL1 0.000000e+00 0.000000e+00 0.000000e+00 3.079390e+01
## WASF2 6.242846e+01 5.469643e+01 3.421535e+01 5.077755e+01
## WDR1 3.418856e+01 4.746531e+01 4.095430e+01 1.610794e+02
## WDR41 1.003195e+01 7.123624e+00 1.645740e+01 0.000000e+00
## WIPF1 1.192175e+01 4.153846e+01 1.596710e+01 2.535619e+01
## XCL1 0.000000e+00 5.266143e+02 0.000000e+00 0.000000e+00
## XCL2 6.292542e+00 2.373878e+03 0.000000e+00 0.000000e+00
## YBX1 1.439178e+03 7.076925e+02 1.177378e+03 4.416247e+02
## YWHAQ 1.211367e+02 3.261467e+02 1.280235e+02 3.728733e+02
## ZAP70 3.587715e+00 1.705450e+02 2.428063e+00 4.091618e+00
## ZCCHC17 4.211349e+01 1.420176e+02 3.556186e+01 1.466200e+02
## ZEB2 6.325769e+00 7.026990e+00 4.054077e+00 0.000000e+00
## ZFP36L2 1.952993e+02 2.952538e+02 2.354933e+02 3.040573e+01
## ZNF593 2.063447e+02 8.166663e+01 2.571223e+02 0.000000e+00
## ABHD11 3.728558e+01 3.026371e+01 1.378697e+01 3.712008e+01
## ABI1 2.234703e+01 9.645245e+01 3.638622e+01 6.168424e+01
## ABRACL 5.947334e+02 6.038601e+02 2.964032e+02 3.824601e+01
## ACAT2 8.595574e+00 8.093559e+01 3.066678e+01 0.000000e+00
## ACOT9 2.042175e+01 8.488368e+00 1.605453e+01 0.000000e+00
## ACRBP 1.007765e+01 0.000000e+00 4.581935e+00 2.481173e+03
## ACTR1A 3.163813e+01 3.480690e+01 8.343906e+01 0.000000e+00
## ADAM28 2.976400e-01 2.377202e+00 1.138708e+01 0.000000e+00
## ADD3 1.296923e+01 6.229476e+01 1.077794e+01 2.287599e+01
## ADK 4.353118e+01 1.066570e+01 2.927468e+01 0.000000e+00
## ADO 1.008399e+00 5.583225e+00 1.606051e+01 0.000000e+00
## AGPAT2 1.469663e+02 4.067778e+01 3.469634e+01 1.186837e+01
## AHNAK 2.876837e+01 2.294932e+01 2.890320e+01 9.368481e+00
## AKIRIN2 1.083893e+02 9.920938e+01 1.880470e+02 5.673503e+02
## AKR1C3 8.037213e-01 3.001357e+02 7.413526e+00 0.000000e+00
## ALDH2 1.348325e+01 1.680929e+00 8.463713e+01 0.000000e+00
## ALDH3B1 2.228277e+01 1.497459e+00 1.351082e+01 0.000000e+00
## ALOX5 6.085731e+01 7.821084e-01 3.771293e+01 0.000000e+00
## ALOX5AP 6.767919e+00 9.225109e+02 3.032069e+02 1.044980e+02
## ANK1 0.000000e+00 0.000000e+00 0.000000e+00 2.228402e+01
## ANXA1 3.306621e+02 6.258894e+02 8.175236e+02 1.341041e+02
## AOAH 6.805963e+01 1.854490e+02 8.163093e+01 6.210391e+01
## AP1S2 1.697815e+02 3.905399e+01 3.666737e+02 6.960316e+01
## APIP 1.169575e+01 2.520887e+01 4.048675e+01 4.167694e+00
## APLP2 5.091119e+01 2.332769e+01 3.634755e+01 2.913421e+00
## AQP3 5.450419e+00 2.710118e+01 1.088372e+01 2.152807e+01
## ARAP1 1.665900e+01 4.697233e+00 2.257422e+00 1.939423e+01
## ARF3 2.229024e+01 2.721280e+01 7.027526e+01 9.902242e+01
## ARF5 3.098852e+02 2.565932e+02 5.280581e+02 3.561978e+02
## ARHGAP21 1.045567e+00 0.000000e+00 1.805159e+00 4.449760e+01
## ARHGAP4 3.933433e+01 2.247407e+01 4.987597e+01 0.000000e+00
## ARHGAP6 0.000000e+00 0.000000e+00 0.000000e+00 8.090225e+01
## ARL6IP4 4.255217e+02 3.976658e+02 3.441905e+02 4.597025e+01
## ARMC10 1.963741e+01 1.038422e+01 4.107747e+01 1.845051e+01
## ARPC1A 1.396798e+02 7.113101e+01 6.519524e+01 0.000000e+00
## ARPC1B 7.226417e+02 3.879928e+02 7.417327e+02 2.690664e+03
## ARPC3 1.871047e+03 8.948931e+02 1.645055e+03 6.827672e+02
## ARPC5L 2.591279e+01 3.812776e+02 8.630246e+01 6.022953e+00
## ARRB1 2.119110e+01 9.671792e+00 2.945329e+00 4.300485e+01
## ARRDC1 5.591086e+01 3.010959e+01 5.793317e+01 5.736902e+00
## ASAH1 9.125909e+01 2.319324e+01 2.281841e+01 2.018842e+02
## ASCL2 7.744701e+01 2.966515e+02 7.261595e+01 0.000000e+00
## ASL 9.123945e+00 1.275764e+01 1.889269e+01 3.605183e+00
## ATG16L2 2.486354e+01 1.959047e+01 6.996394e+00 0.000000e+00
## ATP2B1 2.366335e+01 8.743584e+00 2.756499e+01 4.566388e+00
## ATP6AP2 4.502643e+01 7.085879e+01 3.344285e+01 4.557774e+00
## ATP6V1F 9.825308e+02 9.824541e+02 9.407410e+02 1.301845e+03
## ATXN2 1.693866e+00 2.376617e+00 7.580722e+00 0.000000e+00
## BAG1 3.315921e+01 3.340619e+01 5.877258e+01 4.219171e+01
## BCAP29 1.993884e+01 1.740935e+01 1.631309e+01 3.242078e+01
## BCL7A 5.386413e-01 3.523074e-01 2.778599e+00 0.000000e+00
## BEX2 6.868617e+00 2.639120e+02 1.401098e+01 0.000000e+00
## BIN2 1.809544e+02 4.219517e+02 7.448781e+01 1.897606e+02
## BIRC3 7.609143e+00 4.934416e+00 3.864929e+00 0.000000e+00
## BLK 5.602248e-01 1.348708e+00 1.571393e+01 1.257388e+01
## BLNK 9.945314e-01 2.708502e-01 2.053103e+01 0.000000e+00
## BLOC1S1 6.636534e+02 3.002140e+02 4.257377e+02 1.451270e+02
## BLOC1S2 8.325653e+01 5.252331e+01 8.483311e+01 2.240762e+01
## BLVRA 3.643188e+02 5.483270e+01 9.606582e+01 1.194007e+01
## BMS1 2.919692e+00 2.905473e+00 1.417254e+01 0.000000e+00
## BNIP3L 4.384683e+01 3.079990e+01 3.313309e+01 1.425738e+02
## BPGM 1.522486e+01 1.071708e+02 2.755338e+01 0.000000e+00
## BRI3 2.349483e+02 2.026095e+01 8.050361e+01 9.055495e+01
## BRI3BP 3.015203e+00 8.001616e+00 1.281480e+01 0.000000e+00
## BTK 3.898909e+01 2.848914e+00 3.286147e+01 7.236152e+01
## BZW2 1.899413e+01 2.143918e+01 7.446070e+01 2.303592e+01
## C11orf21 5.575724e+01 4.401769e+01 1.139928e+01 7.056475e+01
## C11orf54 1.114296e+00 6.642641e+00 1.138387e+01 0.000000e+00
## C12orf57 1.010404e+02 2.049217e+02 5.276108e+01 2.197868e+02
## C12orf75 2.134791e+01 3.835937e+02 1.199766e+02 1.872774e+02
## C3AR1 2.788866e+01 2.210658e+01 0.000000e+00 0.000000e+00
## C7orf50 5.827641e+01 3.945294e+01 5.783553e+01 3.400828e+01
## C9orf78 7.923372e+01 1.110705e+02 3.235882e+01 1.229029e+01
## CA2 2.678018e+01 0.000000e+00 3.241017e+01 4.041911e+03
## CALHM2 5.398992e+01 3.910937e+01 3.188046e+01 0.000000e+00
## CAPN1 1.521233e+01 3.019843e+01 6.158074e+00 2.943660e+02
## CAPZA2 5.898398e+01 8.185362e+01 4.316967e+01 1.384297e+02
## CARD16 7.311347e+02 4.490610e+02 3.734897e+02 1.320149e+01
## CARD9 1.760684e+01 1.696245e+00 8.204795e+00 0.000000e+00
## CASP1 3.168814e+02 1.259721e+02 1.677571e+02 0.000000e+00
## CASP4 9.185974e+01 8.564752e+01 5.159601e+01 3.262244e+00
## CAT 1.096368e+02 3.958149e+01 2.599311e+02 0.000000e+00
## CATSPER1 3.296337e+00 6.089783e+01 1.672093e+01 4.005300e+01
## CCDC107 6.957246e+01 3.505389e+02 1.705637e+02 0.000000e+00
## CCDC22 1.844676e+01 3.174152e+01 6.217819e+01 0.000000e+00
## CCDC86 1.058103e+00 4.038483e+00 2.691551e+01 0.000000e+00
## CCDC90B 2.826264e+01 2.152155e+01 1.478994e+01 9.943381e+00
## CD151 1.375983e+01 2.370496e+01 7.198760e+00 3.603963e+02
## CD164 6.155366e+01 1.759358e+02 7.841457e+01 0.000000e+00
## CD27 1.936535e+01 5.443011e+01 1.043186e+01 0.000000e+00
## CD27-AS1 4.568806e+00 1.528448e+01 1.668741e+01 1.635427e+02
## CD36 5.222424e+00 1.056844e+00 4.150302e+00 2.642287e+01
## CD3D 1.904910e+01 1.117310e+02 2.128325e+01 0.000000e+00
## CD3E 4.346537e+01 3.132445e+02 3.989950e+01 1.505931e+02
## CD3G 1.646845e+01 4.986029e+01 0.000000e+00 0.000000e+00
## CD4 4.701974e+01 0.000000e+00 7.566185e+01 0.000000e+00
## CD40LG 1.239697e+00 9.954795e-01 2.196782e+00 0.000000e+00
## CD5 5.593690e+00 2.512732e+00 2.329201e+01 0.000000e+00
## CD6 2.330494e+00 1.981718e+01 6.312331e+00 0.000000e+00
## CD63 2.924757e+02 1.017419e+03 3.814771e+02 6.182816e+02
## CD69 3.372969e+01 2.095684e+02 4.196985e+01 9.187293e+01
## CD72 0.000000e+00 1.303966e+01 3.816434e+01 0.000000e+00
## CD82 7.046417e+00 1.560080e+01 3.777757e+00 1.028288e+03
## CD9 1.516816e+00 0.000000e+00 0.000000e+00 2.522248e+03
## CD99 3.398325e+01 5.662027e+02 1.711800e+02 8.918103e+02
## CDCA7L 4.012596e+00 3.368632e+00 1.108616e+01 0.000000e+00
## CDH23 7.315792e+00 0.000000e+00 2.717149e+00 0.000000e+00
## CDK2AP1 1.285187e+01 0.000000e+00 7.666908e+00 0.000000e+00
## CDKN1B 1.024553e+01 1.026041e+01 1.843985e+01 2.349309e+00
## CDKN1C 2.304819e+02 3.784216e+00 0.000000e+00 2.630185e+01
## CEBPD 4.227094e+02 4.617459e+02 7.024189e+02 0.000000e+00
## CFP 2.362259e+02 5.723123e+00 2.736058e+02 7.741805e+00
## CHCHD2 1.897540e+03 2.272735e+03 2.243590e+03 8.211193e+01
## CHST12 1.897023e+00 5.492454e+01 9.893914e+00 2.716929e+01
## CHST7 4.089465e+01 3.945092e+00 0.000000e+00 0.000000e+00
## CKAP4 2.496099e+01 3.204901e-01 0.000000e+00 0.000000e+00
## CLEC12A 3.788667e+01 2.757957e+00 1.827291e+01 0.000000e+00
## CLEC1B 7.527251e-01 0.000000e+00 0.000000e+00 5.386363e+02
## CLEC4A 4.094681e+01 1.248711e+01 9.220605e+01 1.268447e+02
## CLEC4C 0.000000e+00 0.000000e+00 1.869839e+02 0.000000e+00
## CLEC4E 0.000000e+00 3.844446e+00 0.000000e+00 0.000000e+00
## CLEC7A 6.814289e+01 3.374976e+00 1.885070e+01 0.000000e+00
## CLIC2 0.000000e+00 0.000000e+00 5.934603e+01 0.000000e+00
## CLIC3 1.148078e+01 1.928599e+03 2.523250e+02 0.000000e+00
## CLN8 4.215276e+00 4.207870e+00 1.303561e+01 4.150960e+01
## CLTA 2.267230e+02 1.538081e+02 2.278173e+02 5.195347e+02
## CLU 4.417476e+00 0.000000e+00 2.292638e+00 2.783378e+03
## COMMD6 2.313918e+02 3.046600e+02 3.504159e+02 6.866292e+01
## COMMD9 4.585674e+01 1.759718e+01 4.720459e+01 0.000000e+00
## CORO1B 5.129653e+01 1.834820e+01 4.380736e+01 9.872763e+01
## COX6A1 7.765322e+02 9.566699e+02 8.140526e+02 4.976557e+02
## COX6C 2.346433e+02 3.192116e+02 2.941389e+02 3.112502e+01
## COX8A 1.951328e+03 1.491876e+03 2.211677e+03 8.553684e+02
## CPSF6 6.669473e+00 2.289296e+01 2.841209e+01 0.000000e+00
## CPVL 1.068974e+02 4.432670e+00 3.663048e+02 6.517644e+00
## CSF2RA 7.904706e+00 1.024589e+00 5.491824e+01 0.000000e+00
## CTNNAL1 0.000000e+00 3.207133e+00 0.000000e+00 1.720211e+02
## CTSB 5.655672e+01 8.390532e+00 3.878877e+01 1.731879e+01
## CTSC 5.092477e+01 6.108506e+01 2.302393e+01 6.124995e+00
## CTSD 9.677150e+01 1.733829e+02 2.912773e+01 1.522999e+02
## CTSL 7.340350e+01 1.809537e+00 4.642450e+00 3.400564e+01
## CTSW 2.509271e+01 3.476788e+03 9.044205e+01 1.201927e+01
## CTTN 5.595085e-01 0.000000e+00 0.000000e+00 8.204407e+01
## CUX1 2.020280e+01 3.001034e+00 3.543990e+00 0.000000e+00
## CYB561A3 5.384897e+00 6.539036e+00 2.648498e+01 0.000000e+00
## CYBB 8.538694e+01 8.216344e+00 1.970336e+01 9.048214e+00
## CYC1 1.493765e+02 1.644523e+02 2.783851e+02 3.192471e+01
## DBNL 3.560685e+01 3.926285e+01 3.284326e+01 1.425748e+02
## DCTN3 1.250268e+02 1.528504e+02 7.100963e+01 0.000000e+00
## DDIT4 4.556591e+01 3.004790e+02 2.855061e+02 1.026575e+01
## DENND1A 2.179289e+00 1.581607e+00 8.970076e+00 0.000000e+00
## DERL1 1.821853e+01 4.985372e+01 1.859671e+01 9.011822e+00
## DMTN 0.000000e+00 0.000000e+00 0.000000e+00 6.096714e+02
## DNAJB6 1.382556e+01 2.893471e+01 1.713415e+01 2.404840e+02
## DNAJB9 4.040574e+01 8.657503e+01 4.538865e+01 0.000000e+00
## DNAJC15 1.721678e+02 2.100335e+01 4.452535e+01 8.122099e+00
## DNAJC4 4.721524e+01 3.606764e+01 1.221300e+02 2.053456e+02
## DOK2 2.968101e+02 3.271189e+02 1.694316e+02 3.662358e+02
## DPYSL2 3.092061e+00 0.000000e+00 1.098446e+01 0.000000e+00
## DRAP1 1.089600e+03 9.033953e+02 6.039273e+02 4.007031e+02
## DUSP5 4.859415e+01 9.808321e+00 1.407802e+01 0.000000e+00
## DUSP6 8.282925e+01 2.215638e+01 3.163514e+01 2.749427e+01
## DYNLL1 2.269874e+02 5.299328e+02 1.810148e+02 1.600097e+03
## DYNLT1 1.375141e+02 9.906336e+01 6.594226e+01 7.362973e+00
## EBP 4.241647e+01 2.207124e+02 4.078408e+01 0.000000e+00
## EFNB1 2.362844e+00 9.131819e+00 4.197717e+00 6.614640e+01
## EGFL7 4.481670e+00 0.000000e+00 0.000000e+00 1.707515e+02
## EHBP1L1 1.685631e+01 2.595021e+00 7.991227e+00 3.229297e+00
## EI24 2.996169e+01 7.539506e+01 6.796328e+01 6.228261e+00
## EIF4EBP1 4.124926e+02 9.995798e+01 4.026579e+02 0.000000e+00
## ENDOD1 4.311631e+00 3.778304e+00 3.425175e+00 1.067330e+02
## ENDOG 9.449301e+00 2.604688e+01 7.257878e+01 0.000000e+00
## ENHO 5.123096e+01 0.000000e+00 4.183480e+02 0.000000e+00
## ENKUR 2.025176e+00 0.000000e+00 0.000000e+00 1.334213e+02
## ENTPD1 3.407293e+00 2.125494e+00 5.043518e+00 0.000000e+00
## ENY2 6.300903e+01 4.509114e+01 4.108543e+01 0.000000e+00
## EPSTI1 1.102593e+02 7.481358e+01 1.025839e+02 8.831380e+00
## ERICH1 2.719583e+01 1.506596e+01 7.348941e+00 6.453322e+00
## ERP44 2.556036e+01 2.409599e+01 1.042704e+01 1.529894e+00
## ESAM 9.879882e-01 0.000000e+00 0.000000e+00 4.577504e+02
## ETV6 2.687830e+00 2.549231e+00 1.620438e+01 2.574462e+01
## FABP5 2.623995e+01 3.421651e+01 1.415562e+02 0.000000e+00
## FAM118B 1.127747e+01 5.584233e+00 2.324725e+01 8.229613e+01
## FAM133B 1.595757e+01 4.043447e+01 4.680087e+01 2.497517e+01
## FAM89B 1.269544e+02 6.832532e+01 7.894678e+01 0.000000e+00
## FBP1 9.115254e+01 1.252302e+01 6.511086e+01 0.000000e+00
## FCN1 4.274583e+02 2.056893e+01 1.109632e+02 3.532715e+01
## FDFT1 2.198283e+01 6.357802e+01 8.961044e+01 2.088590e+01
## FERMT3 2.140510e+01 4.562595e+01 2.763559e+01 6.547902e+02
## FGL2 9.353148e+01 1.577096e+01 6.102062e+01 1.342152e+01
## FHL1 1.853308e+00 3.786754e+00 7.136132e+00 3.033840e+02
## FIBP 8.886322e+01 1.078984e+02 1.073988e+02 6.529634e+01
## FKBP11 4.549222e+00 1.104875e+02 1.473467e+01 6.057958e+01
## FKBP15 6.201693e+00 3.897694e+00 1.877311e+00 0.000000e+00
## FLNA 4.310944e+01 7.786756e+01 1.770478e+01 1.857197e+02
## FLT3 0.000000e+00 0.000000e+00 1.766407e+01 0.000000e+00
## FOLR3 8.286701e+00 8.416308e-01 1.093006e+01 0.000000e+00
## FUCA2 3.367503e+01 1.464297e+01 2.694187e+01 0.000000e+00
## FUOM 3.535051e+01 8.641613e+00 2.319905e+02 0.000000e+00
## FUT7 0.000000e+00 1.086177e+01 8.891628e+01 0.000000e+00
## FYN 2.321216e+01 6.006006e+01 2.526521e+01 0.000000e+00
## G6PD 3.435525e+01 5.731456e+01 5.817170e+00 1.322711e+02
## GAPDH 1.670512e+03 2.331060e+03 2.675289e+03 3.174231e+03
## GATA3 4.868821e+00 3.465148e+01 4.060932e+00 0.000000e+00
## GDI2 1.809432e+02 1.212699e+02 4.377270e+02 1.109896e+01
## GFI1B 7.103523e+00 0.000000e+00 0.000000e+00 8.595940e+01
## GHITM 8.570148e+01 1.419533e+02 8.500156e+01 1.025133e+02
## GIMAP4 2.687926e+02 4.645135e+02 1.177919e+02 1.281999e+02
## GIMAP5 3.782423e+01 1.005930e+02 2.348010e+01 0.000000e+00
## GIMAP7 5.916376e+02 1.583678e+03 3.236593e+02 3.582020e+01
## GLA 1.176425e+01 9.472289e+00 0.000000e+00 1.144024e+02
## GLIPR1 6.904144e+01 5.768315e+01 6.536300e+01 0.000000e+00
## GLIPR2 7.730871e+01 2.793068e+02 8.348700e+01 7.117139e+00
## GNB2 2.305321e+02 2.163189e+02 2.450873e+02 1.615368e+01
## GNG11 9.777283e+00 0.000000e+00 0.000000e+00 8.090466e+03
## GNPTAB 3.561284e+00 3.385043e+01 4.336757e+00 1.282345e+01
## GNS 2.523853e+01 9.554249e+00 9.185613e+00 0.000000e+00
## GOLGA7 1.696542e+01 5.814977e+01 1.753954e+01 0.000000e+00
## GPR107 1.227473e+00 0.000000e+00 7.922815e+00 0.000000e+00
## GRINA 1.139184e+02 2.713489e+01 8.438335e+01 1.719483e+01
## GSN 2.054190e+00 1.851606e+01 4.336527e+01 1.791754e+02
## GSTK1 2.564260e+02 2.109443e+02 1.720609e+02 1.483519e+02
## GSTO1 4.785655e+02 3.012371e+02 3.794260e+02 4.642131e+03
## GSTP1 7.162415e+02 9.131779e+02 2.329184e+03 5.030124e+02
## GTF3A 2.355657e+02 3.648536e+02 4.564285e+02 1.195875e+02
## GUSB 1.149217e+02 3.030773e+01 4.766450e+01 7.401455e+01
## HBS1L 2.155899e+00 4.146550e+00 1.161459e+01 0.000000e+00
## HDDC2 1.253582e+01 7.854553e+01 1.876139e+01 0.000000e+00
## HEBP1 3.822047e+01 7.220246e+00 2.951566e+01 0.000000e+00
## HEBP2 6.099572e+00 6.161307e+00 2.365648e+01 8.148912e+00
## HEMGN 0.000000e+00 0.000000e+00 0.000000e+00 2.481328e+02
## HHEX 7.146185e+01 1.366498e+01 6.580604e+01 9.379760e+01
## HNRNPA2B1 7.876492e+01 2.335969e+02 1.250503e+02 9.539977e+01
## HNRNPF 1.451205e+02 2.704209e+02 1.474538e+02 6.692402e+01
## HOTAIRM1 1.669791e+02 5.202048e+01 6.271374e+01 0.000000e+00
## HSP90B1 1.660902e+01 7.122740e+01 2.915405e+01 1.123429e+01
## HSPA5 2.923532e+01 1.192767e+02 4.818302e+01 0.000000e+00
## HSPA8 2.356388e+02 4.655901e+02 2.362352e+02 3.929985e+01
## HVCN1 4.326808e+01 2.699243e+01 1.856371e+01 0.000000e+00
## IDH3G 1.331715e+02 9.354474e+01 1.074752e+02 3.125511e+01
## IDS 3.316702e+01 5.966655e+01 3.466231e+01 3.210910e+01
## IFIT1 4.664370e+01 1.938170e+01 2.165494e+01 1.828295e+01
## IFIT2 3.261071e+01 1.871282e+01 2.980363e+01 0.000000e+00
## IFIT3 5.815709e+01 4.341300e+01 1.100056e+01 6.115680e+01
## IFITM1 5.531261e+02 1.581161e+03 2.799280e+02 2.403523e+02
## IFITM2 5.344590e+03 2.813530e+03 1.480194e+03 4.472003e+02
## IFITM3 1.460676e+03 1.181461e+02 5.379976e+02 9.425542e+01
## IFNG 0.000000e+00 2.408895e+02 0.000000e+00 0.000000e+00
## IL13RA1 0.000000e+00 0.000000e+00 4.116757e+01 0.000000e+00
## IL18 5.906750e+00 3.417532e+00 3.714713e+01 0.000000e+00
## IL2RG 7.833037e+01 5.981174e+02 1.564364e+02 5.377670e+01
## IL3RA 1.218793e+01 5.142123e+00 8.795690e+01 0.000000e+00
## ILK 4.682940e+01 6.232066e+01 3.721915e+01 1.408268e+03
## IMPDH1 7.148550e+01 3.831591e+01 4.775841e+01 3.690707e+01
## INSIG1 8.571432e+01 4.630453e+01 2.954005e+01 1.747918e+02
## IRF5 1.253859e+01 5.944167e+00 1.007188e+01 0.000000e+00
## IRF7 1.374823e+02 1.690553e+02 3.852765e+02 8.324219e+00
## ITGB7 7.064443e+00 1.309676e+02 7.100014e+01 0.000000e+00
## ITM2A 4.056113e+00 6.530797e+01 2.845175e+01 3.180330e+01
## ITM2B 2.188393e+02 2.282997e+02 1.288023e+02 9.332711e+02
## KCNQ1 5.979340e+00 0.000000e+00 2.056437e+01 0.000000e+00
## KCTD12 8.712580e+00 1.208383e+00 1.490089e+01 0.000000e+00
## KLF10 1.923715e+01 3.085523e+01 5.665585e+01 0.000000e+00
## KLF4 1.217838e+02 1.250571e+01 2.028944e+02 0.000000e+00
## KLF6 1.894060e+02 3.407508e+02 2.647156e+02 4.720725e+01
## KLHL42 2.865701e+00 1.522846e-01 9.881916e+00 0.000000e+00
## KLRB1 4.113919e+00 2.721001e+02 0.000000e+00 0.000000e+00
## KLRC1 0.000000e+00 1.039487e+02 0.000000e+00 0.000000e+00
## KLRD1 4.587754e-01 6.960982e+01 8.581372e-01 0.000000e+00
## KLRF1 5.919970e+00 3.928300e+02 0.000000e+00 0.000000e+00
## KLRG1 7.348507e+00 1.823022e+02 2.828950e+01 0.000000e+00
## LAG3 0.000000e+00 4.727107e+00 0.000000e+00 0.000000e+00
## LAMTOR1 1.713081e+02 1.930615e+02 3.575545e+02 2.101801e+03
## LAMTOR4 8.734067e+02 4.042895e+02 4.570877e+02 5.648193e+02
## LAT2 1.505589e+01 8.878144e+01 4.084709e+01 2.470591e+01
## LCN2 0.000000e+00 0.000000e+00 0.000000e+00 3.663515e+02
## LCP1 1.747356e+02 2.438737e+02 1.888345e+02 2.640489e+01
## LDHA 1.753083e+02 2.288807e+02 1.977205e+02 1.220516e+02
## LDHB 1.189237e+02 4.329289e+02 2.847533e+02 1.718359e+02
## LEPROTL1 3.735074e+01 6.531098e+01 2.223783e+01 1.158163e+01
## LFNG 4.497441e+01 2.577574e+01 1.429656e+01 0.000000e+00
## LIMK1 2.755035e+00 5.325827e+00 1.099613e+01 0.000000e+00
## LMO2 3.379421e+01 0.000000e+00 6.126736e+01 7.507893e+00
## LRP1 3.581236e+00 5.508185e-01 6.611498e-01 0.000000e+00
## LSP1 2.801939e+02 2.342164e+02 4.019949e+02 6.016004e+01
## LTA4H 5.177428e+01 2.282140e+01 1.663937e+01 8.808721e+00
## LTV1 2.167399e+01 5.740193e+01 8.049943e+01 0.000000e+00
## LY6E 1.542664e+03 9.110806e+02 7.063783e+02 1.332296e+02
## LYN 1.437617e+02 5.844021e+01 3.343215e+01 8.001366e+01
## LYPD2 3.617934e+02 0.000000e+00 0.000000e+00 0.000000e+00
## LYZ 3.897370e+03 4.691095e+02 1.892507e+04 1.649898e+03
## M6PR 1.559109e+02 1.163548e+02 1.079853e+02 1.474194e+02
## MAD1L1 2.677503e+01 2.389306e+01 1.080271e+01 0.000000e+00
## MAF1 8.142647e+01 2.253947e+02 1.544521e+02 2.551637e+01
## MAP3K11 1.439908e+01 5.684372e+00 1.686327e+01 0.000000e+00
## MAP3K8 3.340059e+01 7.215562e+01 1.827730e+01 6.182217e+00
## MEST 0.000000e+00 2.464688e-01 3.083471e+00 7.508359e+01
## MFSD3 2.026460e+00 1.125599e+01 2.948587e+01 0.000000e+00
## MGST1 1.133907e+00 0.000000e+00 1.507004e+00 0.000000e+00
## MICAL1 1.834802e+01 4.947345e+00 3.150434e+01 0.000000e+00
## MLEC 1.139561e+01 1.356169e+01 3.642803e+01 0.000000e+00
## MOB3B 4.717501e-01 0.000000e+00 4.603774e+00 1.490174e+01
## MPC1 2.270263e+02 1.265291e+02 1.180597e+02 1.411874e+01
## MPEG1 9.598945e+01 7.612701e+00 6.221083e+01 3.385833e+01
## MPP1 5.746805e+01 6.485097e+00 1.415134e+01 9.743449e+02
## MRPL18 3.306026e+02 2.436429e+02 1.916301e+02 3.394404e+02
## MRPL21 9.513674e+01 6.655245e+01 8.768118e+01 4.436687e+01
## MRPL23 1.811976e+02 1.801883e+02 2.167892e+02 5.674693e+01
## MS4A1 1.406936e+01 1.665385e+01 3.673063e+00 2.354054e+01
## MS4A4A 5.839149e+01 0.000000e+00 0.000000e+00 6.689081e+00
## MS4A6A 2.790961e+01 3.992172e+00 1.856900e+02 2.024308e+01
## MS4A7 4.614358e+02 2.361007e+00 2.430950e+01 1.501233e+01
## MSANTD3 1.483200e+00 2.818552e+00 1.223746e+01 6.642796e+01
## MSN 6.122672e+01 6.460794e+01 5.382740e+01 9.811543e+01
## MSRA 3.830057e+01 6.956893e+00 3.097601e+01 2.713590e+01
## MSRB2 4.709571e+01 2.942553e+01 4.836801e+01 0.000000e+00
## MTIF3 1.283088e+02 1.242884e+02 6.871476e+01 2.188106e+02
## MTPN 7.100673e+01 3.210616e+01 3.913120e+01 3.191456e+02
## MTSS1 3.765619e+01 1.796390e+01 2.258022e+00 0.000000e+00
## MTURN 0.000000e+00 6.597926e-01 0.000000e+00 3.446229e+01
## MYC 6.024847e+00 9.996292e+00 9.347417e+00 0.000000e+00
## MYL6 1.296901e+03 1.162415e+03 1.095537e+03 3.440027e+03
## MYO1G 9.342769e+01 6.376772e+01 1.083295e+01 1.217780e+01
## MYOF 8.552146e+00 0.000000e+00 0.000000e+00 0.000000e+00
## NAMPT 1.426373e+01 2.920791e+00 7.632565e+00 9.219326e-01
## NANS 6.186808e+01 3.056922e+01 5.667643e+01 0.000000e+00
## NAP1L1 1.201878e+02 3.473504e+01 4.246364e+01 3.812472e+02
## NCF1 1.577822e+01 5.624766e+00 7.317399e+01 0.000000e+00
## NCKAP1L 2.731830e+01 1.828805e+01 2.414515e+01 2.058983e+00
## NCOA4 7.017735e+01 4.867977e+01 6.301547e+01 2.468065e+03
## NDUFB11 1.297485e+02 1.732163e+02 1.867018e+02 7.637445e+01
## NDUFB2 1.395076e+02 3.264670e+02 1.189626e+02 3.307382e+01
## NEAT1 1.747713e+02 6.190064e+01 7.568360e+01 2.534590e+01
## NECAP1 1.778287e+00 6.391667e+00 2.100039e+01 0.000000e+00
## NEK3 1.529572e+00 8.275321e+00 1.285373e+01 0.000000e+00
## NELL2 1.534110e+00 2.949211e+00 0.000000e+00 3.394670e+00
## NEURL1 2.625087e+01 0.000000e+00 0.000000e+00 4.158129e+00
## NFE2 3.172708e+01 4.514686e+00 0.000000e+00 3.113175e+02
## NINJ1 2.970432e+02 4.086485e+01 1.837003e+01 1.861675e+02
## NIPSNAP3A 6.813772e+01 3.969135e+01 8.456130e+01 8.401414e+00
## NOC4L 1.291814e+01 2.020349e+01 8.249136e+01 3.102386e+01
## NR4A1 2.736146e+01 7.040050e+00 3.524543e+01 2.551523e+01
## NRG1 0.000000e+00 3.094979e-01 7.325282e-01 0.000000e+00
## NRGN 3.940283e+01 6.770104e+00 5.222484e+01 5.816598e+03
## NSMF 1.683622e+01 0.000000e+00 3.114659e+00 0.000000e+00
## NT5C3A 2.637154e+01 3.989164e+01 2.980339e+01 5.597722e+02
## NUCB2 2.347234e+00 1.786827e+01 1.003160e+01 4.325776e+01
## NUP214 1.447818e+01 8.951214e-01 1.086051e+01 0.000000e+00
## OAS1 5.507596e+01 2.564335e+01 2.157307e+01 2.596889e+01
## OAS3 8.519609e+00 4.041921e+00 1.077168e+01 2.409849e+00
## OPTN 5.213869e+00 8.505386e+01 2.908113e+00 0.000000e+00
## ORAI2 1.863674e+00 7.185178e+00 4.070913e+00 0.000000e+00
## OS9 2.008148e+01 3.403786e+01 4.877166e+01 0.000000e+00
## OSTF1 4.840143e+02 7.228626e+02 3.575723e+02 6.159731e+02
## PAK1 2.958913e+01 5.871424e+00 3.693745e+01 7.222341e+00
## PCGF5 3.733603e+01 1.325859e+01 1.686915e+01 7.847519e+00
## PDLIM1 7.307846e+00 5.790335e+01 8.142996e+01 1.420334e+03
## PEBP1 1.222314e+02 3.056641e+02 3.595689e+02 9.714615e+01
## PGAM1 1.673421e+02 2.429568e+02 1.516852e+02 8.560181e+00
## PGK1 2.670425e+02 1.770096e+02 1.393341e+02 1.286188e+02
## PGRMC1 3.016360e+01 6.320054e+01 1.745238e+01 2.277679e+03
## PHF19 2.655278e+01 7.105223e+00 9.268732e+00 3.297370e+00
## PHLDA2 1.364522e+01 0.000000e+00 1.314536e+02 0.000000e+00
## PHPT1 1.628079e+02 2.777440e+02 2.100329e+02 7.775212e+01
## PIK3AP1 3.708756e+01 1.481219e+01 1.058825e+01 0.000000e+00
## PILRA 4.956764e+02 9.963895e+00 9.742495e+01 2.185212e+01
## PKIB 3.713739e+00 0.000000e+00 3.053958e+01 0.000000e+00
## PLBD1 5.593633e+01 2.130197e+00 1.178110e+02 1.834698e+02
## PLEKHF2 1.321268e+01 1.385333e+01 4.913900e+01 5.968394e+00
## PLIN2 4.323926e+01 2.607484e+01 0.000000e+00 4.339026e+00
## PLP2 2.990641e+02 8.016943e+01 5.819794e+02 9.815563e+01
## PLXNC1 9.859002e+00 1.334428e+00 1.286029e+00 0.000000e+00
## PNISR 3.016134e+01 7.934306e+01 4.654637e+01 1.379508e+01
## PNOC 4.724536e+00 3.677601e+00 6.977733e+01 0.000000e+00
## PNPLA4 8.569330e+00 6.959706e+00 2.082505e+01 0.000000e+00
## POLD2 4.914609e+00 6.514914e+00 1.839150e+01 0.000000e+00
## POLD4 3.051470e+02 1.441546e+02 9.858823e+01 7.236498e+01
## POLR2G 1.797502e+02 3.807139e+02 1.975883e+02 4.254080e+02
## POLR2L 4.924322e+02 7.650808e+02 3.746232e+02 2.612420e+02
## POM121 2.545571e+00 1.108106e+01 1.494664e+01 6.355157e+00
## POMP 5.923610e+01 5.957286e+01 9.696481e+01 4.418382e+00
## PON2 0.000000e+00 1.026388e+01 4.470645e+01 0.000000e+00
## POU2AF1 1.341662e+00 3.399319e+00 6.293046e+00 8.957720e+00
## PPA1 3.422825e+01 5.249729e+01 1.868420e+02 2.594487e+01
## PPIA 1.997616e+02 4.616893e+02 3.735618e+02 1.937656e+02
## PPP1CA 3.914597e+02 4.871299e+02 2.600038e+02 1.547999e+02
## PPP1R12A 9.676331e+00 2.871955e+01 1.678284e+01 0.000000e+00
## PPP1R14B 1.395387e+00 1.337694e+01 2.096013e+02 0.000000e+00
## PQBP1 4.725330e+01 5.251138e+01 1.193963e+02 0.000000e+00
## PRDX3 6.195146e+01 7.546948e+01 1.135206e+02 0.000000e+00
## PRDX5 8.421640e+02 1.300767e+03 5.319703e+02 1.805919e+03
## PRF1 1.701702e+01 2.941094e+03 1.700806e+01 7.242528e+01
## PRKAR2B 0.000000e+00 0.000000e+00 1.392616e+00 3.409103e+01
## PRKCQ-AS1 4.666793e+00 2.949214e+01 0.000000e+00 0.000000e+00
## PRR13 3.741582e+02 3.480493e+02 3.257311e+02 8.284124e+01
## PRSS23 3.034628e-01 7.037802e+01 0.000000e+00 0.000000e+00
## PSAP 1.744940e+03 3.290092e+02 5.234415e+02 5.097416e+02
## PSIP1 2.799375e+01 2.795190e+01 2.865981e+01 6.448060e+00
## PSMB1 3.187069e+02 6.148240e+02 4.774968e+02 9.565314e+01
## PSMC3 1.252934e+02 1.844787e+02 8.418581e+01 0.000000e+00
## PSMD9 3.266612e+01 2.866089e+01 2.364363e+01 7.143304e+00
## PSMG3 3.542846e+01 7.490965e+01 9.775798e+01 1.102020e+01
## PTGS1 2.342075e+00 0.000000e+00 7.374397e+00 2.520459e+02
## PTK2 0.000000e+00 0.000000e+00 0.000000e+00 9.303104e+00
## PTMS 5.581491e+00 3.721997e+01 7.134955e+01 1.068973e+02
## PTP4A3 7.317791e+01 2.179216e+00 0.000000e+00 5.889182e+00
## PTPN6 2.307401e+02 1.507166e+02 8.622754e+01 8.790949e+01
## PTPRCAP 1.338296e+02 4.883234e+03 4.780828e+02 1.381879e+02
## PTPRE 9.925534e+00 1.826515e+01 2.971515e+01 0.000000e+00
## QRSL1 1.193510e+01 6.023997e+00 1.187873e+01 0.000000e+00
## RAB18 1.682706e+01 1.381699e+01 1.019348e+01 3.027832e+00
## RAB32 2.042659e+02 0.000000e+00 3.208470e+02 6.713176e+02
## RAC1 1.837468e+02 1.180740e+02 2.289994e+02 2.199960e+02
## RAN 1.499904e+02 2.433796e+02 2.296013e+02 8.917312e+01
## RAP1B 4.877883e+01 1.023682e+02 1.526564e+01 4.436520e+02
## RASSF4 7.143401e+00 6.410855e+00 1.992930e+01 0.000000e+00
## RELT 4.504777e+01 4.210084e+00 2.003625e+01 3.514281e+00
## REPIN1 6.510380e+00 2.613650e+01 4.821000e+01 0.000000e+00
## RGCC 7.673540e+01 5.782858e+01 3.010829e+02 2.668631e+02
## RGS10 5.276302e+02 2.567806e+02 1.029180e+03 9.159271e+03
## RGS3 1.809681e+00 1.991111e+01 2.413241e+00 3.306396e+00
## RHOBTB1 0.000000e+00 0.000000e+00 2.999950e+00 2.220138e+01
## RHOG 8.533831e+02 4.436265e+02 5.974111e+02 3.710385e+01
## RILPL2 2.056855e+02 4.050737e+01 1.701005e+02 2.848339e+02
## RIN1 1.833704e+01 0.000000e+00 0.000000e+00 0.000000e+00
## RMDN1 9.409380e+00 1.117319e+01 2.322783e+01 0.000000e+00
## RNASEH2B 4.754020e+00 8.586632e+00 1.409370e+01 0.000000e+00
## RNASET2 9.499102e+01 2.639522e+01 8.805240e+01 6.545323e+00
## RNH1 1.386519e+02 3.614527e+01 1.680515e+02 8.311547e+01
## RPS6KA4 1.394268e+01 9.172095e+00 7.801151e+01 0.000000e+00
## RPS6KB2 3.011999e+01 2.627944e+01 3.182796e+01 1.077699e+01
## RSU1 3.728910e+01 4.552353e+01 4.314664e+01 4.866447e+02
## RTN3 4.590222e+01 3.538011e+01 1.426761e+01 6.354759e+01
## RWDD1 3.993961e+01 6.719151e+01 4.828045e+01 6.394956e+01
## RXRA 3.528603e+01 5.741067e-01 5.294612e+00 3.149381e+01
## SAMD3 3.345264e+00 1.618643e+02 5.605172e+00 0.000000e+00
## SASH3 6.642228e+01 1.490103e+02 5.788376e+01 4.096620e+01
## SAT1 3.994834e+03 3.087655e+02 1.208154e+03 5.050137e+03
## SBDS 3.900257e+01 1.045612e+02 4.853999e+01 7.748702e+00
## SCGB1C1 2.721045e+01 0.000000e+00 0.000000e+00 2.686949e+03
## SDCBP 1.156285e+02 6.961611e+01 6.982905e+01 7.342544e+01
## SDHAF2 4.462196e+01 1.752072e+02 7.396340e+01 9.672090e+00
## SELPLG 1.594746e+02 2.308173e+02 9.618418e+01 7.769077e+01
## SENCR 7.506612e+00 3.301104e+01 0.000000e+00 5.737600e+02
## SERPINE1 0.000000e+00 0.000000e+00 0.000000e+00 9.904227e+01
## SETDB2 7.474118e+00 5.867962e+00 1.735997e+01 0.000000e+00
## SF3B5 8.876255e+02 2.095107e+03 8.830591e+02 3.920938e+02
## SFT2D1 9.165941e+01 9.347284e+01 8.903140e+01 7.038802e+00
## SH2D1A 6.356720e+00 7.165024e+01 6.398582e+00 0.000000e+00
## SH2D3C 3.561026e+01 2.817166e+01 1.418192e+01 2.426282e+01
## SH3BGRL 7.594580e+02 3.742471e+02 4.147780e+02 1.247568e+03
## SH3BGRL2 0.000000e+00 0.000000e+00 0.000000e+00 1.012015e+02
## SIAE 9.802300e-01 6.645078e-01 0.000000e+00 5.208666e+01
## SIPA1 2.489209e+01 7.770207e+01 1.336225e+01 0.000000e+00
## SIT1 2.024668e+01 1.184585e+01 4.463319e+01 0.000000e+00
## SKAP2 2.603427e+01 5.164472e+01 3.130572e+01 2.094541e+01
## SLC25A3 1.343103e+02 1.461294e+02 1.833415e+02 7.974892e+01
## SLC25A5 9.403436e+02 7.271870e+02 1.426310e+03 2.552109e+02
## SLC25A6 2.078411e+03 1.128805e+03 3.002306e+03 4.884588e+02
## SLC2A3 9.429831e+00 4.551180e+01 2.157529e+01 2.326463e+01
## SLC2A6 7.291586e+01 1.059159e+01 2.511695e+01 0.000000e+00
## SLC31A2 2.632796e+02 1.137842e+01 1.024687e+02 7.178202e+01
## SLC35D2 2.336004e+00 5.330915e+00 2.930133e+00 5.706526e+01
## SMCO4 3.373898e+02 3.495763e+01 1.169653e+02 3.931801e+01
## SMNDC1 9.261923e+00 1.127414e+01 2.648095e+01 0.000000e+00
## SMPDL3A 6.221390e+01 0.000000e+00 0.000000e+00 0.000000e+00
## SMS 1.630724e+02 6.060182e+01 8.407933e+01 1.617567e+02
## SNHG7 2.357705e+01 3.581108e+01 5.992331e+01 1.724045e+01
## SNX10 2.191570e+01 1.421759e+01 1.436803e+01 2.275310e+00
## SNX3 5.806359e+02 6.037241e+02 8.132953e+02 1.336235e+03
## SOD2 5.038591e+01 4.139661e+00 1.005831e+01 1.012921e+02
## SPCS2 1.072928e+02 2.702134e+02 2.650309e+02 7.363719e+01
## SPI1 5.086104e+02 1.585260e+01 2.724491e+02 1.270666e+02
## SPOCK2 3.775778e+00 3.621911e+01 5.079842e+00 3.394670e+00
## SPRYD4 0.000000e+00 2.190944e+00 0.000000e+00 7.838338e+00
## SRGN 3.115738e+03 4.079787e+03 3.818374e+03 1.288256e+04
## SRI 1.249337e+02 1.495940e+02 8.481680e+01 1.029004e+01
## ST14 0.000000e+00 0.000000e+00 1.661318e+01 0.000000e+00
## ST3GAL4 2.718550e+00 2.496485e+01 2.302661e+01 0.000000e+00
## STAC3 4.384125e+01 0.000000e+00 4.882996e+01 0.000000e+00
## STAT2 1.531144e+01 5.845986e+00 9.618713e+00 0.000000e+00
## STIP1 1.194074e+01 5.034003e+01 1.752371e+01 0.000000e+00
## STK32C 1.042375e+01 1.109873e+01 3.176293e+01 0.000000e+00
## STK38L 4.334930e+00 4.858673e+00 2.228118e+01 0.000000e+00
## STOM 1.644598e+01 1.034656e+02 8.157564e+00 2.236504e+02
## STRBP 3.991661e-01 5.451954e-01 0.000000e+00 0.000000e+00
## STX11 9.099523e+01 2.082059e+01 1.925058e+01 2.758062e+02
## STX7 1.730905e+01 1.014551e+01 1.537833e+01 1.326836e+00
## SUSD3 9.922886e+00 1.786823e+01 1.509534e+02 1.016242e+02
## SWAP70 1.938896e+01 6.959855e-01 5.336465e+00 3.023201e+00
## SWI5 1.283994e+01 2.736082e+01 3.867230e+01 4.023220e+02
## SYK 2.688216e+01 1.849331e+01 6.118787e+01 3.358091e+01
## SYNE1 7.717075e-01 5.367686e+00 3.838458e-01 0.000000e+00
## SYPL1 2.741560e+01 1.797571e+01 4.260573e+01 1.818514e+02
## SYTL4 0.000000e+00 0.000000e+00 0.000000e+00 6.481369e+01
## TAF10 1.176097e+01 1.463757e+01 2.580639e+01 1.076958e+01
## TALDO1 3.174640e+02 1.416632e+02 2.950205e+02 4.786471e+03
## TBC1D10C 8.635271e+01 1.809255e+02 2.788612e+01 0.000000e+00
## TBL2 6.355105e+00 7.210426e+00 1.419812e+01 0.000000e+00
## TBXAS1 8.776114e+01 2.756394e+01 1.646654e+01 2.521840e+01
## TCEAL8 3.658328e+01 2.079329e+02 1.226875e+02 1.968962e+02
## TCF7L2 3.395333e+01 4.406385e+00 3.938002e+00 0.000000e+00
## TCIRG1 8.442961e+01 4.300038e+01 3.278132e+01 8.961671e+01
## TDRP 1.019836e+00 0.000000e+00 0.000000e+00 1.342526e+02
## TES 2.343322e+01 7.888529e+01 2.080745e+01 3.852608e+01
## TESC 2.810149e+02 3.263824e+01 4.570045e+01 6.640539e+01
## TIMM8B 1.957361e+02 2.271793e+02 1.657725e+02 5.374632e+01
## TIMP1 2.417649e+03 3.096270e+02 5.379900e+02 2.726961e+03
## TJP2 1.833942e+00 0.000000e+00 0.000000e+00 2.260327e+01
## TLN1 2.126920e+01 2.090504e+01 2.597367e+01 1.304792e+02
## TMEM123 8.833782e+01 8.963817e+01 9.012695e+01 6.694929e+01
## TMEM134 3.905900e+01 3.450817e+01 2.401706e+01 3.690234e+00
## TMEM140 9.332943e+00 2.481887e+02 1.226570e+01 5.165328e+02
## TMEM141 1.356680e+02 4.027059e+02 8.932638e+01 0.000000e+00
## TMEM176A 1.658737e+01 0.000000e+00 0.000000e+00 0.000000e+00
## TMEM176B 1.022898e+02 9.138421e+00 0.000000e+00 1.203924e+01
## TMEM179B 2.210850e+02 2.443411e+02 2.000501e+02 0.000000e+00
## TMEM218 4.476578e+00 4.352285e+00 1.382698e+01 0.000000e+00
## TMEM243 3.784797e+01 8.694750e+01 6.877028e+01 0.000000e+00
## TMPO 1.704357e+01 1.020983e+01 1.125914e+01 1.730752e+01
## TNFAIP3 1.800416e+01 2.637454e+01 2.475710e+01 1.742796e+01
## TNFRSF1A 4.936922e+01 4.845846e+01 3.569202e+01 1.034143e+01
## TNNI2 2.165778e+02 0.000000e+00 4.218312e+01 3.996849e+01
## TOLLIP 2.496236e+01 1.720705e+01 2.058045e+01 1.223682e+02
## TOMM7 6.878393e+02 6.334094e+02 6.425220e+02 3.087993e+02
## TPD52 2.131929e+00 6.762317e+00 9.908626e+00 7.726323e+00
## TPI1 6.631606e+02 5.783617e+02 5.153694e+02 2.293708e+02
## TPP1 2.529451e+01 2.357029e+01 5.456373e+01 0.000000e+00
## TSC22D1 4.201867e+00 3.474252e+00 0.000000e+00 9.542765e+02
## TSC22D3 3.502052e+02 3.339548e+02 3.015912e+02 8.292634e+02
## TSPAN13 0.000000e+00 5.139658e+00 8.442452e+01 1.329320e+02
## TSPAN14 1.388393e+01 1.052506e+01 3.902166e+00 1.184832e+00
## TSPAN18 0.000000e+00 0.000000e+00 0.000000e+00 1.764223e+01
## TSPAN33 7.909803e-01 7.883163e-01 6.353325e+00 5.644753e+01
## TTYH3 2.041577e+01 0.000000e+00 2.384110e+00 3.553335e+00
## TUBA1A 3.558872e+02 1.849072e+02 3.120875e+02 7.508242e+01
## TUBA1B 4.241571e+02 4.717324e+02 6.176964e+02 6.964907e+02
## TUBA1C 8.370786e+00 2.908714e+01 2.116392e+01 3.789305e+02
## TXN 1.403138e+02 3.876524e+02 8.225013e+02 1.051029e+02
## UBC 8.917034e+02 1.158211e+03 8.220335e+02 1.101285e+03
## UBE2A 3.426845e+01 3.542297e+01 4.843904e+01 5.211360e+01
## UBE2D1 9.626241e+01 2.783500e+01 4.876785e+01 6.247948e+00
## UBE2J1 5.367556e+01 3.152046e+01 4.577349e+01 5.973783e+01
## UBE2L6 3.871307e+02 3.858293e+02 3.834308e+02 2.108910e+01
## UBE2R2 3.952272e+01 2.114688e+01 3.188216e+01 0.000000e+00
## UBL4A 7.029957e+00 3.413646e+01 2.316749e+01 4.863382e+02
## UCHL3 7.123970e+01 5.839105e+01 1.435309e+02 1.150416e+01
## UCP2 3.617823e+02 8.738122e+02 7.066925e+02 3.128436e+02
## UNC93B1 9.415023e+01 7.843363e+00 8.363780e+01 0.000000e+00
## UPF3B 1.198596e+01 6.618866e+01 9.039682e+01 0.000000e+00
## UPP1 6.215909e+01 6.204997e+01 1.573550e+01 1.368301e+01
## UTRN 2.329871e+01 9.379050e+00 8.098490e+00 0.000000e+00
## VCL 4.386831e+00 4.644991e+00 1.647075e+01 2.185636e+01
## VIM 7.650505e+02 2.208125e+02 1.746572e+03 9.960969e+01
## VMA21 4.020176e+01 2.781393e+01 1.128848e+01 1.227104e+01
## VPS29 1.728809e+02 1.005000e+02 1.388255e+02 6.754538e+00
## WAS 1.198630e+02 1.033725e+02 1.113238e+02 7.061270e+01
## WNT11 0.000000e+00 0.000000e+00 0.000000e+00 4.474922e+01
## XPNPEP1 4.898675e+00 1.373465e+01 1.608823e+01 2.866852e+02
## YBX3 1.773815e+01 1.818237e+00 2.262709e+01 9.785122e+00
## YIF1A 6.709365e+01 5.581512e+01 9.865148e+01 0.000000e+00
## YTHDF3 4.986282e+00 6.774409e+00 1.780857e+01 1.016846e+01
## YWHAZ 6.434568e+01 6.805464e+01 5.189384e+01 9.465841e+01
## ZBTB16 7.039333e-01 1.066121e+01 0.000000e+00 0.000000e+00
## ZCCHC7 1.280302e+01 2.632494e+01 2.980528e+01 0.000000e+00
## ZFAND5 7.530076e+01 1.696667e+01 3.950795e+01 6.283266e+01
## ZNF385A 4.515971e+01 0.000000e+00 9.039355e+01 0.000000e+00
## ZNF398 2.090001e+00 1.789456e+00 9.874592e+00 0.000000e+00
## ZNF467 1.225657e+01 8.135413e-01 6.161374e+01 0.000000e+00
## ZNF503 0.000000e+00 0.000000e+00 2.176952e+01 0.000000e+00
## ZNF703 6.418320e+01 6.319518e+00 9.251545e+00 6.556954e+00
## ZNF706 1.079055e+02 4.456742e+01 8.581490e+01 6.161820e+01
## ZNF789 1.702396e+00 2.158956e+00 1.218672e+01 0.000000e+00
## ZNHIT1 8.581727e+01 9.545770e+01 1.157999e+02 5.623918e+01
## ZYX 1.060431e+02 4.837037e+01 7.972883e+01 2.005853e+02
## AATK 2.683773e-01 0.000000e+00 0.000000e+00 1.300619e+01
## ABCC3 3.057263e+00 0.000000e+00 0.000000e+00 6.040835e+01
## ABHD17A 4.951247e+00 7.324228e+01 8.166657e+00 1.016846e+01
## ABI3 5.719992e+02 4.192444e+02 2.557291e+02 1.913984e+01
## ACAA2 2.278369e+01 6.870048e+01 1.511067e+01 5.414549e+01
## ACAP1 3.798026e+00 7.306369e+01 2.926510e+01 0.000000e+00
## ACO2 1.578325e+01 1.170727e+01 9.643583e+00 1.840743e+00
## ACTG1 1.050390e+03 1.824575e+03 1.643521e+03 1.963692e+03
## ACTN1 2.050818e+00 9.269683e-01 2.951782e+00 1.860158e+02
## ADA 4.334054e+01 2.878165e+01 1.365905e+01 7.142271e+00
## ADAP2 2.626654e+01 0.000000e+00 1.323127e+01 2.994356e+01
## ADCY7 1.351852e+01 1.158740e+01 6.846707e+00 0.000000e+00
## AHSA1 3.463757e+01 1.075721e+02 4.534591e+01 5.963200e+01
## ALDOA 6.990779e+02 4.794381e+02 5.285472e+02 1.105008e+03
## ALOX12 0.000000e+00 0.000000e+00 0.000000e+00 4.120457e+02
## ANAPC11 9.825605e+01 1.306207e+02 1.015344e+02 1.330559e+02
## ANKRD12 1.778294e+01 3.094400e+01 2.450056e+01 6.887454e+00
## ANKRD9 9.919309e-01 8.648909e-01 0.000000e+00 9.202701e+01
## ANXA2 1.857860e+02 1.103848e+02 2.392033e+02 3.045390e+01
## AP2A1 3.634342e+01 1.067318e+01 9.634589e+00 9.686833e+00
## AP2S1 9.381307e+02 3.842889e+02 8.754372e+02 1.394197e+03
## APEX1 8.750131e+01 1.579536e+02 4.104212e+02 0.000000e+00
## APH1B 2.192958e+01 5.463353e+00 0.000000e+00 3.994646e+00
## APMAP 2.181702e+01 6.527659e+02 3.795966e+01 8.192581e+01
## APOBEC3A 4.956952e+02 5.225775e+00 1.747046e+01 5.583707e+01
## APOBEC3B 1.841830e+02 3.075633e+00 1.903469e+01 0.000000e+00
## APOBEC3G 5.268011e+01 2.319515e+02 4.722592e+01 0.000000e+00
## APOBR 1.333389e+01 5.193942e+01 3.117628e+01 1.652643e+01
## ARG2 7.101498e+00 0.000000e+00 0.000000e+00 2.089694e+02
## ARHGAP27 1.681503e+01 7.907547e+00 1.382542e+01 0.000000e+00
## ARHGDIA 1.631265e+02 2.578648e+02 1.024696e+02 6.626486e+01
## ARHGEF40 3.383427e+01 0.000000e+00 1.737912e+01 2.653510e+00
## ARL6IP1 3.887639e+01 1.893605e+02 6.765078e+01 1.717015e+02
## ARRB2 2.307777e+02 1.084828e+02 1.219041e+02 5.097974e+00
## ASGR1 2.208743e+01 7.594278e+00 2.425886e+01 5.801190e+00
## ATP2A3 1.451193e+00 1.665886e+01 1.166790e+01 1.481160e+02
## ATP6V0D1 6.044295e+01 3.936605e+01 3.600191e+01 8.915888e+01
## ATP9A 0.000000e+00 0.000000e+00 0.000000e+00 7.409777e+01
## BAX 2.924737e+02 3.437433e+02 1.379364e+02 5.782223e+01
## BCL2A1 1.874806e+02 1.455285e+01 1.296757e+01 0.000000e+00
## BID 2.775785e+02 4.803770e+01 1.485739e+02 6.556681e+01
## BLVRB 1.381711e+02 4.705705e+01 2.954924e+02 1.788390e+02
## BSG 8.142669e+01 2.063207e+02 6.935485e+01 1.441320e+02
## BST2 9.342130e+02 1.646480e+03 1.124770e+03 3.003911e+02
## C15orf39 2.099704e+01 5.760878e+00 1.160140e+01 1.079184e+01
## C15orf48 4.912252e+00 7.167796e+00 9.105431e+01 0.000000e+00
## C16orf74 6.290978e+00 7.456706e-01 3.191251e+01 0.000000e+00
## C17orf49 3.656112e+01 7.875731e+01 1.313129e+02 2.384635e+01
## C19orf33 0.000000e+00 0.000000e+00 2.269240e+01 2.560865e+03
## C19orf38 3.677072e+02 7.570339e-01 1.167938e+02 2.801915e+01
## C5AR1 2.159515e+02 1.127208e+01 8.993083e+00 5.255535e+01
## CACTIN 1.409197e+00 1.277291e+00 0.000000e+00 3.317072e+01
## CALM1 1.600615e+02 4.714324e+02 1.630720e+02 2.635798e+02
## CALM3 1.564558e+02 1.324865e+02 4.091396e+01 1.388881e+03
## CALR 1.127052e+02 4.135887e+02 1.575050e+02 6.642885e+01
## CAPNS1 1.036582e+02 6.258777e+01 1.994727e+02 1.300396e+02
## CARHSP1 2.148249e+01 6.701137e+01 2.339461e+01 5.173405e+01
## CARS2 1.042665e+01 3.689591e+00 6.301617e+00 0.000000e+00
## CBFA2T3 8.028370e+00 0.000000e+00 2.015902e+01 0.000000e+00
## CBR1 9.870741e+01 9.490291e+01 1.099041e+02 8.030151e+01
## CCL3 1.049153e+02 1.542832e+03 3.178854e+02 0.000000e+00
## CCL4 2.846876e+01 2.899852e+03 3.128971e+01 0.000000e+00
## CCL5 9.713998e+01 6.424383e+03 1.694639e+02 1.442208e+04
## CCR7 1.393042e+01 7.564346e+00 1.728856e+01 0.000000e+00
## CD19 6.802099e+00 1.928926e+00 6.550033e+00 0.000000e+00
## CD22 2.058630e+00 3.751659e-01 1.249893e+01 0.000000e+00
## CD2BP2 3.753030e+01 8.624296e+01 3.269008e+01 1.171175e+01
## CD300A 1.339717e+02 2.953275e+02 9.762933e+01 0.000000e+00
## CD300C 1.370258e+02 3.854044e+01 1.201015e+02 0.000000e+00
## CD300E 7.613503e+01 0.000000e+00 1.260678e+01 0.000000e+00
## CD300LB 2.907255e+00 0.000000e+00 2.309030e+01 0.000000e+00
## CD300LF 1.384639e+02 4.794936e+00 4.154926e+01 0.000000e+00
## CD320 0.000000e+00 1.139939e+02 0.000000e+00 2.749812e+01
## CD33 3.075980e+01 0.000000e+00 8.180571e+01 0.000000e+00
## CD37 5.165050e+02 3.573659e+02 3.968375e+02 1.580951e+02
## CD40 1.027663e+01 6.768590e+00 1.892012e+01 0.000000e+00
## CD68 9.408226e+02 2.329189e+01 2.492984e+02 5.656207e+02
## CD7 1.183504e+01 1.236954e+03 5.585024e+00 0.000000e+00
## CD79A 3.121626e+01 5.976013e+01 1.260532e+02 1.617772e+01
## CD79B 4.032887e+02 2.029091e+02 7.356558e+01 3.069580e+01
## CDIP1 3.693621e+01 1.788875e+01 1.385473e+01 2.235791e+02
## CDKN2D 3.267373e+02 3.920835e+02 1.138591e+02 4.064980e+03
## CEACAM3 2.970801e+01 0.000000e+00 0.000000e+00 6.712340e+01
## CEBPA 4.853823e+01 3.538303e+00 4.056775e+01 5.153065e+01
## CEBPB 1.692739e+03 2.126924e+02 2.230432e+02 1.911146e+02
## CEBPG 3.285043e+01 2.475550e+01 1.134377e+01 1.545074e+01
## CENPN 2.074784e+00 5.936719e+00 1.498350e+01 0.000000e+00
## CERK 2.328917e+01 8.753720e+00 9.371541e+00 0.000000e+00
## CFD 1.581603e+03 1.979927e+01 6.895051e+01 5.621091e+02
## CHAF1A 9.772769e+00 3.957175e+00 2.414399e+01 0.000000e+00
## CHCHD10 7.810490e+02 7.744775e+01 2.880534e+02 2.143588e+02
## CHEK2 2.338316e+00 2.743832e+00 1.263638e+01 0.000000e+00
## CHMP1B 1.718218e+02 1.095573e+02 1.957408e+02 3.259680e+02
## CHMP4B 2.558075e+02 1.122336e+02 2.219823e+02 2.026105e+02
## CHMP6 2.909983e+01 4.001528e+01 0.000000e+00 5.615510e+02
## CIITA 4.426473e-01 1.597736e+00 5.020892e+00 0.000000e+00
## CINP 2.582941e+01 1.483857e+01 1.638694e+01 0.000000e+00
## CISD3 5.112452e+01 1.901205e+02 5.376495e+01 4.019770e+01
## CKB 1.723965e+02 6.797331e+00 1.530970e+01 0.000000e+00
## CLDN5 1.133865e+00 0.000000e+00 0.000000e+00 9.598378e+02
## CLEC10A 0.000000e+00 6.819101e-01 4.933574e+02 0.000000e+00
## CLPP 1.501452e+01 6.458774e+01 3.692447e+01 0.000000e+00
## CMTM3 5.294085e+01 7.654706e+01 1.290941e+02 0.000000e+00
## CMTM5 1.336091e+00 0.000000e+00 0.000000e+00 1.140216e+03
## COMT 3.769927e+01 3.699562e+01 3.465698e+01 1.817126e+01
## COPE 2.929673e+02 3.470007e+02 3.944830e+02 1.598620e+02
## CORO1A 6.157736e+02 6.868473e+02 6.215624e+02 1.975371e+02
## COTL1 1.599152e+03 8.436964e+01 7.775188e+02 7.210576e+02
## COX5A 5.286563e+02 5.681701e+02 5.176200e+02 1.109195e+02
## COX6B1 1.197606e+03 7.898276e+02 1.021016e+03 5.276912e+02
## CPPED1 8.808410e+01 1.129646e+01 4.224928e+01 0.000000e+00
## CRELD2 1.215395e+01 5.891915e+01 1.906066e+01 0.000000e+00
## CRIP1 3.386551e+02 2.433265e+02 2.227109e+02 1.295919e+02
## CRIP2 7.364724e-01 0.000000e+00 0.000000e+00 0.000000e+00
## CSK 1.038256e+02 1.376381e+02 6.430744e+01 4.669035e+01
## CST3 3.300244e+03 1.239009e+02 6.610516e+03 1.329106e+03
## CST7 3.967195e+01 7.431000e+03 1.103458e+02 0.000000e+00
## CSTB 6.445032e+02 2.030684e+02 4.323913e+02 1.186848e+02
## CTNNBL1 4.872638e+01 4.564119e+01 1.705107e+01 4.835062e+01
## CTSA 3.080952e+01 3.263720e+01 1.661451e+01 6.568563e+02
## CTSH 2.710355e+01 2.738375e+00 5.935813e+01 1.632454e+01
## CTSZ 2.375549e+01 1.320557e+00 2.295914e+01 1.486477e+01
## CXCL16 1.376586e+02 0.000000e+00 4.971557e+01 1.660205e+02
## CYB5B 8.280060e+00 5.304983e+01 1.187048e+01 0.000000e+00
## CYBA 1.675800e+03 1.574621e+03 1.597950e+03 4.325207e+02
## CYLD 5.395980e+00 9.063111e+00 4.554503e+01 0.000000e+00
## CYTH4 5.619172e+01 4.573709e+01 4.746369e+01 2.197593e+01
## DAAM1 1.156099e+00 4.270976e+00 0.000000e+00 4.164001e+01
## DDX5 1.515210e+02 3.406466e+02 1.925584e+02 1.358511e+02
## DEDD2 9.244086e+01 8.631793e+01 3.583194e+01 8.264104e+00
## DENND6B 1.702591e+01 6.409802e+00 9.275722e+00 0.000000e+00
## DESI1 2.283538e+01 6.906083e+00 1.624066e+01 4.420179e+00
## DET1 2.609138e+00 0.000000e+00 1.194787e+01 0.000000e+00
## DHRS4L2 4.383856e+01 5.155752e+01 2.679854e+02 0.000000e+00
## DHRS7 1.110284e+02 4.216614e+02 1.097301e+02 9.153239e+01
## DNAJB1 2.544530e+01 3.224948e+01 2.386912e+01 0.000000e+00
## DNASE2 4.489932e+01 5.626784e+01 8.311147e+00 0.000000e+00
## DPEP2 7.193971e+01 2.529958e+01 0.000000e+00 6.215797e+00
## DSTN 8.115452e+01 1.491007e+02 5.703231e+01 2.555415e+02
## DTD2 4.575463e+00 2.292863e+00 3.070221e+01 0.000000e+00
## DUSP3 4.583185e+00 3.307615e+00 1.810039e+01 0.000000e+00
## EGLN3 0.000000e+00 1.448893e+00 0.000000e+00 2.031592e+01
## EIF3G 2.263356e+02 5.026841e+02 4.319586e+02 9.479239e+01
## EIF4A1 4.086520e+02 4.952917e+02 6.217532e+02 3.832656e+01
## EIF6 2.623805e+02 2.717655e+02 3.796481e+02 1.584648e+02
## EMC7 2.058992e+02 2.126118e+02 1.344268e+02 2.994884e+01
## EMC8 2.873591e+01 2.278056e+01 4.300739e+01 9.958394e+01
## EMP3 2.158448e+03 2.044646e+03 1.051872e+03 4.519882e+02
## EPN1 1.080516e+01 1.110286e+01 7.268910e+00 1.234268e+00
## ERCC1 4.294058e+01 2.564749e+01 6.098627e+01 2.014141e+01
## ETFA 2.466062e+01 2.881182e+01 2.727547e+01 1.770689e+02
## ETHE1 7.690126e+01 1.356558e+02 1.367041e+02 1.367475e+01
## EVI2B 3.386918e+02 1.158595e+02 1.831752e+02 1.054966e+01
## EVL 6.366477e+01 1.628701e+02 3.871267e+01 1.071249e+01
## FAH 1.421535e+00 4.804146e+00 1.567807e+00 8.062336e+01
## FAM110A 2.017305e+02 2.792424e+01 4.877378e+01 3.095457e+02
## FAM118A 4.273531e+00 6.649951e+00 2.101951e+01 0.000000e+00
## FAM32A 1.429635e+02 1.152905e+02 6.252466e+01 1.137332e+02
## FCER2 7.720141e+00 1.447274e+01 5.264271e+00 8.871015e+00
## FCGRT 1.680640e+02 1.303668e+01 2.699829e+02 5.990758e+01
## FCHO1 9.047219e+00 1.377549e+00 1.546694e+01 0.000000e+00
## FES 1.307082e+01 1.897712e+01 4.139447e+01 0.000000e+00
## FKBP1A 3.857533e+02 1.222943e+02 2.377695e+02 8.643260e+02
## FKBP3 3.962894e+01 1.165027e+02 3.736719e+02 0.000000e+00
## FLT3LG 1.747217e+01 2.446294e+01 2.109220e+01 0.000000e+00
## FMNL1 6.393755e+01 6.465485e+01 5.737054e+01 2.065421e+01
## FN3K 0.000000e+00 0.000000e+00 0.000000e+00 9.893311e+01
## FNTB 9.944554e-01 0.000000e+00 8.803665e+00 1.235367e+02
## FOSB 1.453581e+02 8.422637e+01 2.099825e+02 1.313549e+02
## FPR1 1.608684e+02 4.477154e-01 3.647156e+01 8.417677e+00
## FXYD5 6.073563e+02 5.164274e+02 5.730953e+02 8.929766e+02
## G6PC3 6.289297e+00 2.556829e+00 1.058453e+01 0.000000e+00
## GAA 1.437725e+01 3.183662e+00 1.444804e+01 0.000000e+00
## GABARAP 1.069072e+03 3.436980e+02 1.209004e+03 7.887414e+02
## GAS2L1 1.947353e+00 2.600515e-01 9.092087e+00 3.409209e+02
## GAS6 6.182421e-01 0.000000e+00 2.242348e+01 0.000000e+00
## GAS7 4.613743e+00 6.854699e+00 7.721401e+00 0.000000e+00
## GCH1 6.826675e+01 4.285098e+01 2.620546e+01 0.000000e+00
## GCHFR 2.926847e+01 3.113872e+02 4.451758e+01 0.000000e+00
## GLOD4 1.706454e+01 4.491373e+01 2.489505e+01 0.000000e+00
## GMFG 1.753274e+03 1.589205e+03 1.019168e+03 5.587209e+02
## GMIP 2.874647e+01 1.618209e+01 4.707946e+00 7.214254e+01
## GNA15 1.122487e+01 0.000000e+00 6.089168e+01 7.414084e+01
## GNAS 2.236639e+01 2.247305e+01 3.030453e+01 5.870917e+01
## GNB5 1.145932e+00 3.336993e+00 2.734378e+00 3.895671e+01
## GNG2 4.210309e+01 8.826571e+01 1.111680e+01 3.408506e+00
## GNG7 1.563390e+00 2.334142e+00 1.838337e+01 0.000000e+00
## GNGT2 3.681102e+01 8.047185e+01 4.640057e+01 0.000000e+00
## GP1BA 0.000000e+00 0.000000e+00 0.000000e+00 6.678689e+02
## GP6 0.000000e+00 0.000000e+00 0.000000e+00 5.534077e+01
## GPR18 7.603889e+00 1.163140e+01 6.895462e+00 0.000000e+00
## GPR183 7.518391e+00 1.872637e+01 2.022169e+02 0.000000e+00
## GPX4 6.547834e+02 3.916995e+02 8.329124e+02 1.850990e+03
## GRAP2 3.527777e+00 2.870977e+01 5.149473e+00 1.087214e+03
## GRB2 6.643509e+01 5.552340e+01 5.804537e+01 9.674499e+01
## GRN 1.768940e+02 2.916816e+01 4.941195e+02 1.257817e+02
## GUCD1 6.843850e+00 2.014246e+01 2.551449e+00 0.000000e+00
## GZMB 4.050230e+01 9.688523e+03 1.641775e+03 2.198777e+02
## GZMH 3.341193e+01 3.045490e+03 2.308441e+01 0.000000e+00
## GZMM 1.053563e+01 2.683729e+03 7.633001e+01 0.000000e+00
## HAUS1 4.148346e+01 4.545500e+01 9.861301e+01 0.000000e+00
## HCK 4.096486e+02 6.646444e+00 9.051304e+01 1.410500e+01
## HCST 1.106697e+03 5.540800e+03 6.764536e+02 1.055291e+03
## HERPUD1 7.294717e+01 3.479593e+01 1.630547e+02 4.295310e+00
## HEXA 1.777948e+00 1.811818e+01 1.633311e+01 0.000000e+00
## HEXIM2 8.567963e+00 9.825364e+00 0.000000e+00 1.976768e+02
## HMG20B 3.166664e+01 3.318834e+01 2.950593e+01 6.740152e+01
## HMGN1 7.034339e+01 3.176217e+02 2.195796e+02 4.048661e+02
## HMOX1 2.908129e+02 1.463512e+01 6.374201e+00 9.428820e+00
## HMOX2 2.838246e+01 1.858754e+02 8.551782e+01 0.000000e+00
## HSBP1 8.150390e+01 2.418806e+01 2.495606e+01 1.737264e+01
## HSCB 4.751104e+01 2.949456e+01 1.187457e+02 4.310055e+01
## HSH2D 3.380426e+01 1.321716e+02 7.978742e+01 0.000000e+00
## ICAM2 1.651612e+02 2.312039e+02 5.847489e+01 4.783585e+02
## ICAM3 3.024864e+02 3.105912e+02 2.133816e+02 7.321009e+01
## ICAM4 1.050092e+02 0.000000e+00 2.139321e+01 0.000000e+00
## ID1 6.705966e+00 1.584957e+01 5.341958e+02 0.000000e+00
## IDH2 1.306245e+02 1.643439e+02 1.406614e+02 2.054150e+02
## IDH3A 8.331062e+00 1.075794e+01 2.353478e+01 0.000000e+00
## IER2 2.991155e+02 6.584067e+02 4.374087e+02 8.706152e+01
## IFI27L2 1.581642e+02 1.461979e+02 1.380969e+02 3.462311e+02
## IFI30 6.600366e+02 7.182747e-01 3.213601e+02 0.000000e+00
## IFI35 1.975647e+02 2.340653e+02 7.449588e+01 0.000000e+00
## IFNGR2 1.834655e+02 1.961074e+01 1.213767e+02 8.563552e+00
## IGFLR1 3.852349e+01 5.043836e+01 1.141334e+02 0.000000e+00
## IGLL5 5.495575e+01 3.202356e+01 2.498207e+01 4.680225e+01
## IGSF6 1.013108e+02 3.575185e+00 1.762565e+02 3.238776e+01
## IL10RB 2.407581e+01 1.754889e+01 1.418190e+01 2.709390e+00
## IL12RB1 1.828660e+01 7.291193e+01 6.432449e+00 0.000000e+00
## IL17RA 1.095044e+01 7.297014e+00 9.961692e+00 4.202356e+01
## IL2RB 1.990868e+00 1.180782e+02 6.003315e+00 0.000000e+00
## IL32 3.806036e+01 1.065455e+03 8.881017e+01 6.775587e+00
## IL4R 1.235022e+01 6.706355e+00 5.069366e+00 0.000000e+00
## IMPA2 1.901583e+01 4.258465e-01 4.593670e+01 0.000000e+00
## IQGAP1 1.268833e+01 1.803034e+01 1.203912e+01 1.889533e+00
## IRF8 1.741957e+01 1.177358e+01 1.133454e+02 0.000000e+00
## ISG20 3.133787e+01 1.543578e+02 5.341479e+01 3.489321e+01
## ISOC2 1.482318e+01 2.707146e+01 4.232880e+01 1.421693e+01
## ITGA2B 0.000000e+00 4.918743e-01 0.000000e+00 9.900796e+02
## ITGAL 3.003002e+01 4.264739e+01 1.157910e+01 1.886999e+01
## ITGAM 2.473593e+00 3.000944e+01 1.436033e+01 3.822584e+00
## ITGAX 2.044120e+01 5.020709e+00 6.074508e+00 3.369719e+01
## ITGB2 2.253407e+02 4.434362e+02 1.444312e+02 1.122957e+02
## ITGB3 0.000000e+00 0.000000e+00 0.000000e+00 1.790889e+02
## ITPK1 1.813622e+01 8.992338e+00 6.151922e+00 1.653731e+01
## JOSD2 1.094987e+02 3.988022e+01 1.672787e+02 0.000000e+00
## JUND 9.147790e+01 7.377428e+01 1.072372e+02 6.497481e+01
## KIAA0513 5.022409e+00 0.000000e+00 0.000000e+00 8.068230e+01
## KIAA0930 1.468658e+01 1.403798e-01 1.772547e+01 0.000000e+00
## KIF16B 1.557802e+00 1.495473e-01 1.138530e+01 0.000000e+00
## KIFC3 0.000000e+00 1.816648e+00 0.000000e+00 3.028895e+01
## KIR2DL3 0.000000e+00 1.307861e+02 0.000000e+00 0.000000e+00
## KIR3DL2 2.157522e+00 1.722876e+02 0.000000e+00 0.000000e+00
## KPNA2 5.347852e+00 1.936502e+01 2.594524e+01 0.000000e+00
## KSR1 2.015283e+00 7.178096e-01 6.280990e+00 0.000000e+00
## LACTB 4.217695e+01 1.568450e+01 3.662887e+01 5.519893e+00
## LAIR1 2.204794e+01 4.208666e+01 1.119204e+01 0.000000e+00
## LAMP1 3.048851e+01 1.396725e+02 1.487285e+01 1.848117e+01
## LASP1 2.612793e+01 3.504247e+01 1.614097e+00 4.571574e+01
## LAT 1.182299e+01 1.016804e+02 3.333900e+00 1.237489e+02
## LDLRAD4 1.801375e+00 2.350065e-01 2.559889e+00 0.000000e+00
## LGALS1 6.983351e+03 2.757409e+03 6.583444e+03 1.745908e+03
## LGALS2 3.484619e+02 3.467459e+01 2.590370e+03 2.695999e+02
## LGALS3 5.650256e+02 5.531606e+01 3.319903e+02 7.674789e+01
## LGALS9 1.120059e+02 3.644299e+01 9.329956e+01 2.345664e+01
## LGMN 4.154323e+00 3.031261e-01 4.445664e+01 0.000000e+00
## LILRA1 4.328327e+01 4.215209e+00 2.673200e+01 0.000000e+00
## LILRA2 3.256397e+01 1.693935e+00 2.547591e+01 0.000000e+00
## LILRA4 0.000000e+00 0.000000e+00 2.792970e+02 0.000000e+00
## LILRA5 4.874176e+02 1.849233e+01 6.886435e+01 0.000000e+00
## LILRB1 8.810479e+01 5.578380e+00 7.917602e+00 3.951810e+00
## LILRB2 1.777621e+02 4.739583e+00 6.024011e+01 1.138668e+01
## LILRB3 5.079532e+01 4.295468e+00 3.755675e+01 3.005577e+01
## LILRB4 3.123460e+01 0.000000e+00 7.610277e+01 0.000000e+00
## LIMD2 3.018280e+02 4.663910e+02 3.461858e+02 6.427282e+01
## LINC00528 1.042439e+01 7.486963e+00 6.097103e+01 0.000000e+00
## LINC00926 2.935846e+00 1.802832e+00 0.000000e+00 0.000000e+00
## LITAF 2.936634e+01 2.675087e+02 8.935541e+01 4.298707e+00
## LRRC25 3.835849e+02 3.072536e+00 8.479240e+01 4.479369e+01
## LYL1 1.348993e+02 3.087952e+00 5.623678e+01 3.430288e+02
## LYSMD2 2.284046e+02 1.186884e+02 1.515086e+02 1.160238e+01
## MAFB 1.481765e+02 8.441382e+00 3.908865e+01 1.283143e+01
## MAFG 3.183867e+00 1.621184e+00 2.674136e+01 3.752068e+01
## MAP2K3 9.226689e+01 8.511446e+01 4.956564e+01 3.386576e+01
## MAP3K7CL 3.885153e+00 1.862807e+00 0.000000e+00 5.153347e+02
## MAP4K1 5.258113e+00 3.219697e+01 5.367029e+01 1.599087e+01
## MAPK1 2.829380e+01 5.251641e+01 9.366530e+00 0.000000e+00
## MATK 0.000000e+00 1.809960e+02 0.000000e+00 0.000000e+00
## MAX 2.122375e+01 4.599400e+01 3.533557e+01 5.772682e+02
## MBD2 2.752941e+01 3.466832e+00 2.425417e+00 0.000000e+00
## MBOAT7 3.123747e+01 0.000000e+00 3.874425e+01 0.000000e+00
## MBP 5.347229e+00 2.036007e+01 7.122157e+00 3.482572e+00
## METTL9 2.503635e+01 6.439371e+01 3.888966e+01 1.214373e+01
## MIDN 5.588384e+01 3.912907e+01 4.280030e+01 3.876724e+01
## MIEN1 1.458834e+02 2.584817e+02 7.938700e+01 1.067396e+01
## MIF 6.271633e+02 1.145873e+03 9.509322e+02 7.845324e+02
## MILR1 1.223141e+01 0.000000e+00 4.342387e+01 0.000000e+00
## MIR142 3.342336e+03 5.340904e+03 3.669671e+03 9.996717e+02
## MIR24-2 2.758491e+03 1.913522e+03 4.697924e+03 2.978474e+02
## MIS18BP1 2.111449e+01 1.504658e+01 1.835975e+01 0.000000e+00
## MLH3 0.000000e+00 4.035954e+00 1.787735e+00 1.363895e+02
## MLX 6.887338e+01 3.858579e+01 4.540851e+01 0.000000e+00
## MMD 9.443810e+00 3.446565e+00 1.406316e+01 1.505148e+03
## MOB3A 3.680582e+01 3.882901e+01 2.976211e+01 5.942571e+01
## MPST 5.816249e+00 1.757264e+01 2.700612e+01 1.863498e+02
## MRPL27 6.954265e+01 5.290948e+01 7.075114e+01 0.000000e+00
## MRPL54 3.540535e+02 3.624480e+02 3.271706e+02 4.414794e+01
## MRPS6 2.117539e+01 1.833939e+02 7.270412e+01 0.000000e+00
## MSRB1 1.167945e+02 3.776671e+01 9.773925e+01 3.176986e+02
## MT-ND3 9.325991e+02 5.830779e+02 6.813020e+02 1.256813e+02
## MT2A 1.868624e+03 8.722246e+02 5.988454e+02 3.884397e+02
## MTHFS 7.402357e+01 3.664047e+01 3.135706e+01 1.436595e+01
## MVD 1.129279e+01 4.840073e+01 9.067177e+00 0.000000e+00
## MX2 1.393192e+01 1.154391e+01 1.274986e+01 0.000000e+00
## MYADM 7.777775e+01 3.169179e+01 9.336810e+01 0.000000e+00
## MYL12A 7.912330e+02 2.227190e+03 1.262007e+03 5.449918e+03
## MYL12B 1.125641e+03 1.353293e+03 7.770483e+02 6.485657e+02
## MYL9 3.387461e+00 8.430315e+00 0.000000e+00 2.106142e+03
## MYO1F 6.994538e+01 1.365378e+02 4.551062e+01 2.923327e+01
## MYO9B 1.402149e+01 9.648832e+00 7.787261e+00 1.128299e+01
## NAGA 5.575270e+01 1.206834e+01 3.519639e+01 0.000000e+00
## NAPA 3.637627e+01 4.707931e+01 5.678845e+01 5.816634e+01
## NCF4 3.496856e+01 9.421949e+00 5.454795e+01 9.710969e+00
## NDRG2 1.067145e+00 0.000000e+00 3.458269e+01 0.000000e+00
## NDUFA11 1.655626e+02 1.439681e+02 2.184389e+02 6.436661e+01
## NDUFA13 4.084586e+02 2.913254e+02 3.110550e+02 6.234200e+01
## NDUFB1 1.134219e+02 1.216096e+02 9.608301e+01 0.000000e+00
## NDUFB10 4.682812e+02 6.265568e+02 3.809173e+02 8.116449e+01
## NDUFB7 8.777647e+02 1.486923e+03 5.414831e+02 9.416803e+02
## NDUFS7 7.515010e+01 6.273415e+01 6.435947e+01 8.748468e+00
## NDUFV3 2.293426e+01 1.576406e+01 3.088258e+01 0.000000e+00
## NFKBIA 2.822149e+02 2.327347e+02 3.416852e+02 1.793627e+02
## NKG7 4.131218e+02 3.604152e+04 1.781963e+02 2.135716e+02
## NLK 1.090817e+00 0.000000e+00 0.000000e+00 5.138081e+01
## NME1 2.498415e+01 3.057982e+01 5.074338e+01 0.000000e+00
## NME1-NME2 5.369395e+01 4.244552e+01 1.534452e+02 1.752043e+01
## NOP10 5.467648e+02 5.395054e+02 2.593730e+02 1.523678e+02
## NOSIP 1.158850e+02 1.171020e+02 5.876686e+01 4.856569e+00
## NPC2 1.188968e+03 1.229541e+02 8.629962e+02 3.033318e+02
## NT5C 1.095912e+02 2.178165e+02 1.729559e+02 4.087946e+01
## NT5M 0.000000e+00 0.000000e+00 0.000000e+00 2.920355e+02
## OAZ1 3.400884e+03 9.711708e+02 1.824027e+03 8.635283e+03
## OAZ2 8.819497e+01 3.340514e+01 3.457683e+01 0.000000e+00
## OGFOD3 7.891961e+00 1.488957e+01 1.972790e+01 0.000000e+00
## OLIG1 5.029775e+01 0.000000e+00 0.000000e+00 0.000000e+00
## OSCAR 1.748253e+02 0.000000e+00 3.541207e+01 0.000000e+00
## OXA1L 5.485015e+01 4.835585e+01 9.584495e+01 0.000000e+00
## P2RX1 3.763126e+01 0.000000e+00 7.314683e+00 2.278883e+02
## P2RX5 1.312741e+00 5.305034e+00 1.819107e+01 1.554657e+01
## P4HB 2.448874e+01 6.325510e+01 2.537018e+01 3.154770e+01
## PACSIN2 2.835618e+01 7.385613e+00 2.709782e+01 1.591519e+02
## PARVB 8.489470e+00 3.639667e+00 2.609663e+01 4.914968e+02
## PARVG 3.368358e+01 1.806750e+01 3.282326e+01 2.029423e+01
## PATL2 4.292829e+00 4.964918e+01 0.000000e+00 0.000000e+00
## PCK2 2.111179e+01 5.387484e+00 1.683414e+01 4.710048e+01
## PCNA 5.519662e+01 1.988113e+02 6.131779e+01 4.466729e+01
## PCP2 0.000000e+00 0.000000e+00 0.000000e+00 7.178039e+02
## PDCD5 1.345863e+02 1.119324e+02 7.499277e+01 5.878819e+01
## PDIA3 3.177378e+01 1.286996e+02 4.860359e+01 2.387098e+01
## PDXK 1.330265e+01 5.232873e+00 2.362637e+00 0.000000e+00
## PER1 1.158073e+01 2.861971e+00 2.150273e+01 0.000000e+00
## PFKL 2.500880e+01 1.533354e+01 2.609141e+01 2.984492e+01
## PGLS 3.476251e+02 1.149035e+02 3.231730e+02 2.296781e+01
## PIK3IP1 2.446054e+01 6.255284e+01 5.227471e+00 2.872560e+01
## PITPNC1 1.175205e+01 3.500676e+01 2.242769e+00 2.783264e+00
## PKIG 2.588709e+00 3.436689e+00 0.000000e+00 2.700570e+02
## PKM 7.950879e+01 7.032615e+01 1.074078e+02 5.729118e+02
## PKN1 5.634628e+01 6.058167e+01 3.707519e+01 4.703192e+00
## PLA2G4C 4.362506e-01 0.000000e+00 0.000000e+00 6.231024e+01
## PLAGL2 1.609511e+01 5.171443e+01 6.786104e+00 6.900898e+01
## PLAUR 1.193841e+02 1.298371e+01 3.238380e+01 9.756724e+00
## PLD4 4.068654e+01 4.383574e+00 2.795136e+02 0.000000e+00
## PLEKHF1 0.000000e+00 2.754070e+02 5.614765e+00 0.000000e+00
## PLIN3 2.005361e+01 1.267160e+01 4.520210e+01 7.938247e+00
## PLXNB2 2.097495e+01 2.456970e+00 1.088497e+01 2.990353e+00
## PMAIP1 1.100811e+02 1.583409e+02 1.775959e+02 0.000000e+00
## PNMA1 1.021850e+01 1.145081e+01 5.883379e+00 6.053441e+02
## POLR3K 1.253589e+02 6.262172e+01 2.876883e+01 3.353034e+01
## POU2F2 7.590453e+01 5.524248e+00 2.959067e+01 2.196426e+01
## PPCDC 2.951506e+01 7.413108e+00 2.411116e+01 2.250962e+01
## PPDPF 2.717209e+03 1.928869e+03 1.696345e+03 5.680851e+03
## PPIB 1.215646e+02 3.838872e+02 3.110723e+02 2.283851e+02
## PPM1F 1.422007e+01 1.854186e+00 7.853898e+00 0.000000e+00
## PPM1N 8.855264e+01 3.663959e+00 1.074000e+01 5.744474e+01
## PPP1R14A 0.000000e+00 1.127814e+00 1.914369e+02 3.291764e+02
## PPP2R3C 4.377691e+01 4.956619e+01 4.993310e+01 0.000000e+00
## PPP2R5C 8.902854e+00 4.623267e+01 1.070440e+01 0.000000e+00
## PPP4C 1.865712e+02 1.861697e+02 1.427091e+02 2.386702e+01
## PRAM1 7.618816e+01 1.054663e+01 2.714100e+01 0.000000e+00
## PRDX2 3.982038e+01 2.108096e+02 7.345218e+01 3.081033e+01
## PRKCB 4.125233e+01 1.754546e+01 1.346126e+01 4.510897e+01
## PRKCH 2.466849e+00 4.985405e+01 8.086400e+00 0.000000e+00
## PRMT2 3.093318e+01 1.076553e+02 2.808653e+01 8.040212e+01
## PRR14L 2.718305e+00 0.000000e+00 5.811771e+00 9.508710e+00
## PRR5 1.244332e+01 4.442399e+01 0.000000e+00 0.000000e+00
## PSENEN 1.727817e+02 2.864532e+02 1.955475e+02 0.000000e+00
## PSMA4 3.742630e+01 4.128043e+01 3.362697e+01 1.206907e+01
## PSMA7 8.222545e+02 1.263284e+03 6.300063e+02 2.836715e+02
## PSMB10 3.244988e+02 8.906401e+02 3.729202e+02 1.337199e+01
## PSMB3 8.038827e+02 6.311445e+02 7.154143e+02 1.795783e+02
## PSMC4 1.320139e+02 2.032990e+02 1.024457e+02 1.411988e+02
## PSMC5 6.467347e+01 1.090210e+02 1.293542e+02 9.195545e+01
## PSMD8 9.093011e+01 1.725440e+02 1.540113e+02 2.461255e+01
## PSME1 9.973551e+02 1.728166e+03 1.103592e+03 2.687020e+02
## PSME2 4.779662e+02 6.394002e+02 4.170517e+02 8.383388e+01
## PTGDR 0.000000e+00 2.685973e+02 2.072181e+01 3.480984e+01
## PTGER2 6.506992e+01 2.169047e+02 1.791680e+01 0.000000e+00
## PVALB 0.000000e+00 0.000000e+00 0.000000e+00 7.355792e+02
## PYCARD 1.399947e+03 4.234953e+02 8.700531e+02 1.614834e+02
## PYGL 1.276937e+01 0.000000e+00 1.219117e+01 1.677054e+02
## RAB11A 2.779031e+01 4.148936e+01 4.566577e+01 3.950923e+02
## RAB22A 1.618856e+00 1.468707e+01 1.694326e+01 0.000000e+00
## RAB27B 1.061095e+00 7.119844e+00 0.000000e+00 6.460211e+01
## RAB31 3.125067e+01 2.969882e-01 2.790959e+01 1.937553e+01
## RAB34 1.206658e+01 9.598079e-01 2.105520e+01 0.000000e+00
## RAB4B 3.943995e+01 4.560885e+01 4.701332e+01 0.000000e+00
## RAB5C 2.170784e+02 1.978013e+02 2.011127e+02 2.206247e+02
## RAB8A 1.128622e+02 9.405859e+01 5.357045e+01 2.701252e+02
## RABGGTA 1.206811e+01 3.916166e+01 3.505226e+01 6.363143e+00
## RAC2 5.403351e+02 1.378233e+03 3.747409e+02 1.662298e+01
## RALY 6.849361e+01 8.217062e+01 4.102729e+01 1.412937e+01
## RASAL3 3.717607e+01 8.130501e+01 1.027757e+01 0.000000e+00
## RASSF2 1.900026e+01 1.350126e+00 1.053927e+01 0.000000e+00
## RBM39 5.020585e+01 8.182170e+01 4.893888e+01 5.447518e+00
## RBM42 4.648040e+01 8.313863e+01 1.036902e+02 1.971247e+01
## RBX1 2.611667e+02 1.656541e+02 1.380209e+02 4.778593e+02
## RCN2 5.171410e+00 4.347840e+01 4.677585e+00 0.000000e+00
## RDH11 1.352147e+01 3.001690e+01 9.404804e+00 1.936769e+02
## RETN 2.862224e+02 0.000000e+00 9.773222e+01 0.000000e+00
## RGL4 2.763778e+01 1.689369e+01 0.000000e+00 0.000000e+00
## RGS19 3.859219e+02 1.834740e+02 2.917859e+02 1.072133e+01
## RIN3 2.577953e+01 2.660287e+01 1.931130e+01 5.012185e+00
## RNASE2 6.947233e+00 0.000000e+00 3.891624e+01 0.000000e+00
## RNASE6 1.863680e+01 4.396885e+01 8.542469e+02 0.000000e+00
## RNF125 7.048417e+00 4.039636e+01 5.338599e+00 0.000000e+00
## RNF126 5.219447e+01 1.615552e+02 5.045717e+01 6.298627e+00
## RNF135 7.680998e+01 3.545377e+01 2.872565e+01 9.680703e+00
## RNF167 5.444028e+01 1.643865e+02 4.238692e+01 0.000000e+00
## RNF213 1.976401e+01 2.357513e+01 8.190313e+00 2.296833e+01
## RORA 1.502678e+00 1.584259e+01 7.995337e-01 0.000000e+00
## RPL36AL 2.456240e+03 3.478194e+03 2.033142e+03 5.121775e+02
## RPS21 1.321302e+03 1.429237e+03 2.254573e+03 7.779731e+02
## RRAS 8.691587e+01 2.419965e+01 0.000000e+00 0.000000e+00
## RRP7A 3.820170e+01 2.962097e+01 1.445164e+01 0.000000e+00
## RSL1D1 4.583083e+01 5.418753e+01 7.641703e+01 1.822271e+01
## RTN1 6.151092e-01 0.000000e+00 1.214146e+01 0.000000e+00
## S100B 2.936956e+01 4.443305e+02 0.000000e+00 0.000000e+00
## S1PR5 0.000000e+00 2.581943e+02 0.000000e+00 0.000000e+00
## SAMD14 0.000000e+00 0.000000e+00 0.000000e+00 6.195113e+01
## SAMHD1 4.126338e+01 4.453202e+01 7.654171e+01 2.072697e+01
## SAT2 8.599316e+01 1.229573e+02 1.949538e+02 2.646727e+01
## SCAMP4 7.636620e+00 1.101727e+01 2.201083e+01 0.000000e+00
## SCIMP 6.511568e+01 8.372930e+00 5.641669e+01 4.177852e+01
## SCN1B 0.000000e+00 0.000000e+00 2.141541e+00 6.038545e+01
## SCO2 2.346513e+02 9.038550e+01 1.797874e+02 0.000000e+00
## SCPEP1 9.837461e+01 2.058495e+01 2.886882e+01 6.957320e+01
## SDF2L1 1.764339e+02 4.397151e+02 1.644547e+02 7.970828e+01
## SEC11A 1.444836e+02 1.302858e+02 1.218418e+02 3.959006e+00
## SEC14L1 1.782044e+01 8.215139e+00 7.950255e+00 1.234485e+01
## SEC14L5 0.000000e+00 0.000000e+00 0.000000e+00 5.469639e+01
## SECTM1 4.463583e+01 3.975418e-01 2.852432e+01 0.000000e+00
## SERF2 3.952590e+02 5.217786e+02 3.545761e+02 8.775947e+02
## SERPINA1 5.885613e+02 9.768752e+00 5.827818e+01 4.383277e+01
## SERPINF1 0.000000e+00 0.000000e+00 2.322004e+02 0.000000e+00
## SERPINF2 0.000000e+00 0.000000e+00 3.845169e+01 0.000000e+00
## SERTAD3 8.447027e+01 5.749756e+01 6.496303e+01 0.000000e+00
## SETD3 7.267266e+00 4.125568e+01 8.157290e+00 2.165221e+01
## SH3BP1 9.972229e+01 4.335564e+01 2.605914e+01 0.000000e+00
## SHKBP1 8.765017e+01 8.559677e+01 4.875251e+01 0.000000e+00
## SIGLEC10 1.339862e+02 4.340795e+00 1.255476e+01 5.927715e+00
## SIGLEC9 1.758331e+01 7.276675e+00 3.673630e+01 0.000000e+00
## SIK1 1.788805e+01 5.770725e+00 8.579394e+00 0.000000e+00
## SIRPB1 1.581546e+01 1.248986e+00 1.081458e+01 0.000000e+00
## SKAP1 1.012678e+01 2.011672e+02 1.238808e+01 8.217256e+00
## SLC16A3 3.923249e+01 1.869664e+01 1.071181e+01 1.215768e+01
## SLC24A4 5.525938e+00 0.000000e+00 2.144043e+00 1.471399e+00
## SLC35B1 1.209423e+01 2.134048e+01 4.189466e+01 0.000000e+00
## SLC39A3 9.441590e+00 1.886857e+01 6.469204e+01 1.655667e+02
## SLC43A2 1.126092e+01 0.000000e+00 2.675206e+00 1.058825e+01
## SLC44A2 4.219282e+01 4.904257e+01 0.000000e+00 9.012879e+01
## SLC7A7 1.304026e+02 0.000000e+00 1.844353e+01 4.050458e+00
## SLIRP 7.830179e+01 6.415119e+01 6.392610e+01 0.000000e+00
## SMARCB1 7.228095e+00 1.586099e+01 1.660656e+01 0.000000e+00
## SMIM5 0.000000e+00 0.000000e+00 1.449385e+01 1.229261e+02
## SMOX 0.000000e+00 0.000000e+00 1.041348e+01 1.582406e+02
## SMPD3 0.000000e+00 0.000000e+00 1.622032e+01 0.000000e+00
## SNAP23 5.269936e+01 4.445423e+01 3.975485e+01 3.160171e+02
## SNAP29 4.224344e+01 2.820372e+01 3.824956e+01 1.765917e+01
## SNF8 5.080996e+01 9.990923e+01 4.441203e+01 3.291106e+01
## SNN 4.438022e+01 0.000000e+00 1.863728e+01 1.161201e+03
## SNRNP25 6.203235e+01 6.024679e+01 6.941431e+01 2.795192e+01
## SNRPD1 7.328640e+01 8.836486e+01 1.162504e+02 5.981788e+01
## SNRPD2 3.663405e+02 6.511981e+02 5.514381e+02 3.750344e+02
## SNX29P2 1.202469e+01 2.397399e+01 6.315386e+01 0.000000e+00
## SNX5 5.339212e+01 2.576509e+01 2.149841e+01 0.000000e+00
## SOCS3 3.456701e+01 5.405640e+00 4.993309e+01 0.000000e+00
## SOD1 5.835130e+02 3.436992e+02 2.676179e+02 2.580064e+02
## SPECC1 7.360691e-01 1.488921e+00 5.779735e+00 5.284655e+00
## SPG21 9.122100e+01 3.813907e+01 7.396794e+01 1.091783e+01
## SPHK1 9.224767e-01 0.000000e+00 0.000000e+00 1.684248e+02
## SPIB 1.848747e+00 1.350361e+00 4.926817e+01 0.000000e+00
## SPINT1 6.187926e+00 0.000000e+00 2.405853e+01 0.000000e+00
## SPINT2 2.262448e+01 2.205401e+00 1.563277e+02 4.023549e+02
## SPN 6.755210e+01 6.993955e+01 3.448760e+01 2.945389e+00
## SPPL2A 6.612905e+00 3.076589e+01 8.323805e-01 1.402675e+00
## SPTSSA 4.758287e+01 3.059293e+01 3.941800e+01 1.358716e+02
## SRSF5 2.930254e+02 2.807465e+02 2.611565e+02 8.117550e+00
## ST13 1.215949e+02 1.370059e+02 1.456101e+02 7.655118e+01
## STX10 1.544342e+02 9.813066e+01 7.158829e+01 2.621917e+02
## STXBP2 3.126069e+02 4.130449e+01 5.957497e+01 2.597903e+02
## SULF2 1.093973e+01 2.192642e+00 4.231454e+01 0.000000e+00
## SULT1A1 1.216519e+02 5.553373e+00 1.060758e+01 1.100081e+02
## SUMO3 1.071574e+02 1.407511e+02 1.951689e+02 2.567201e+02
## SUN2 2.354914e+01 9.502209e+01 1.988234e+01 1.864664e+01
## SUPT4H1 3.978236e+02 2.974256e+02 2.894870e+02 4.168723e+02
## SYNE2 1.025257e+00 5.429061e+00 8.905857e-01 0.000000e+00
## SYNGR1 4.390331e-01 3.616311e+01 2.104625e+01 1.617988e+01
## SYNGR2 1.058118e+02 2.377754e+01 8.572007e+01 1.610185e+01
## TAX1BP3 1.112520e+02 5.423484e+01 7.474068e+01 8.183701e+02
## TBCB 1.973027e+02 2.301713e+02 1.267837e+02 6.477366e+01
## TBX21 8.145829e+00 2.049984e+02 0.000000e+00 0.000000e+00
## TBXA2R 0.000000e+00 3.272486e+00 0.000000e+00 9.266617e+01
## TCF4 4.753395e-01 1.354582e+00 2.539194e+00 0.000000e+00
## TCL1A 1.610293e+01 1.433336e+01 1.511893e+02 0.000000e+00
## TECR 1.769047e+01 9.226815e+01 3.127982e+01 0.000000e+00
## TGFB1 1.243819e+02 1.523623e+02 5.544141e+01 4.757678e+02
## TGFB1I1 0.000000e+00 0.000000e+00 0.000000e+00 1.086532e+02
## TIMM13 2.076959e+02 1.991323e+02 1.828610e+02 8.838899e+01
## TMC6 1.826070e+01 1.454836e+01 6.882946e+00 1.685232e+00
## TMED1 5.075565e+01 1.083610e+02 5.138987e+01 0.000000e+00
## TMED3 8.981982e+00 1.069553e+01 4.548200e+00 4.304670e+01
## TMEM11 7.418417e+01 4.708430e+01 2.103066e+01 0.000000e+00
## TMEM205 5.517378e+01 1.107944e+02 1.193745e+02 6.991803e+01
## TMEM219 1.605091e+02 1.577245e+02 2.480516e+02 7.597231e+02
## TMEM91 6.295431e+00 1.429407e+01 0.000000e+00 5.770795e+02
## TMIGD2 3.489842e+00 2.135351e+02 1.732697e+01 0.000000e+00
## TMSB4Y 0.000000e+00 8.649581e-01 1.342410e+01 7.935706e+01
## TNFAIP2 3.037566e+01 8.291107e-01 5.079044e+01 0.000000e+00
## TNFSF13B 1.878096e+02 1.461088e+01 1.954084e+02 4.381537e+01
## TNNC2 5.759438e+00 0.000000e+00 0.000000e+00 9.965887e+01
## TOMM40 1.068617e+01 2.155041e+01 4.311101e+01 6.022953e+00
## TOP1 7.932443e+00 1.187510e+01 1.779794e+01 0.000000e+00
## TPM1 2.857633e+00 1.955265e+00 0.000000e+00 2.484270e+02
## TPM4 7.479241e+00 2.107027e+01 3.194734e+01 6.485684e+02
## TPPP3 1.382403e+02 1.945105e+00 5.684688e+01 0.000000e+00
## TPST2 1.740704e+01 1.521116e+02 5.701640e+00 3.535445e+02
## TPTEP1 3.808824e+02 4.589782e+01 0.000000e+00 4.419874e+03
## TRADD 6.757017e+01 6.938341e+01 1.178455e+02 0.000000e+00
## TRAPPC1 6.568704e+02 1.200433e+03 9.258700e+02 3.046143e+03
## TRAPPC5 1.943369e+01 5.467109e+00 1.514818e+01 1.868294e+01
## TSEN34 3.427056e+01 3.875859e+01 4.155289e+01 1.224733e+02
## TSEN54 8.033561e+00 5.661631e+01 1.337081e+01 1.391081e+01
## TSPAN3 7.578177e+00 1.417232e+01 5.305242e+00 0.000000e+00
## TSPO 1.075952e+03 4.029382e+02 9.672523e+02 2.857524e+02
## TTC38 6.120835e+00 2.795142e+02 3.134047e+00 0.000000e+00
## TTC39C 5.575712e+00 1.182498e+01 1.394224e+01 0.000000e+00
## TUBA8 0.000000e+00 0.000000e+00 3.712822e+00 8.705171e+01
## TUBB1 5.753868e+00 8.202374e-01 1.142869e+01 4.534127e+03
## TUBB6 7.686589e+00 5.512644e+00 4.097702e+01 5.006330e+01
## TXN2 1.755058e+02 1.405829e+02 1.703398e+02 2.635498e+01
## TXNDC17 5.642375e+01 6.087752e+01 6.381175e+01 0.000000e+00
## TXNL1 2.376513e+01 2.512453e+01 1.783234e+01 1.924658e+00
## TYMP 8.841717e+02 1.104874e+02 5.451407e+02 1.515552e+02
## TYROBP 1.092864e+04 5.241060e+03 5.589318e+03 1.242289e+03
## UBB 9.395865e+02 3.905807e+03 2.026854e+03 2.607946e+03
## UBL5 3.988893e+02 3.993777e+02 4.350564e+02 5.912528e+02
## UBL7 6.893457e+01 5.567879e+01 1.511356e+02 0.000000e+00
## UNC119 8.280571e+01 1.724320e+01 3.736990e+01 4.271681e+00
## UPK3A 3.549359e+01 0.000000e+00 1.910086e+02 0.000000e+00
## UQCR10 6.034891e+02 7.023136e+02 6.963377e+02 3.375015e+02
## UQCRFS1 1.547811e+02 1.831931e+02 1.818403e+02 2.113693e+01
## USF2 2.144013e+01 2.417700e+01 4.793956e+01 2.637991e+02
## USP10 3.409157e+00 8.004069e+00 1.158710e+01 1.299323e+02
## UTY 2.883096e+00 3.248512e+00 1.274920e+01 0.000000e+00
## VASP 3.038238e+02 1.849147e+02 1.449294e+02 3.475350e+02
## VMO1 2.186181e+02 0.000000e+00 4.457527e+01 0.000000e+00
## VMP1 5.307014e+01 1.734883e+01 1.647233e+01 2.174068e+00
## VPREB3 1.118415e+01 6.118778e+00 2.691069e+01 0.000000e+00
## VPS35 1.511183e+01 1.009973e+01 7.763942e+00 4.467864e+00
## VSTM1 1.336427e+02 5.498518e+00 0.000000e+00 0.000000e+00
## WDR83OS 9.443426e+02 8.951510e+02 8.193001e+02 4.259044e+02
## WIPI1 2.747084e+01 1.986390e+01 0.000000e+00 1.586544e+02
## WSB1 5.097473e+01 4.801881e+01 3.581269e+01 0.000000e+00
## XBP1 5.779637e+01 5.379354e+02 1.120263e+02 0.000000e+00
## YIF1B 1.746511e+01 2.041915e+01 6.793490e+01 1.053947e+02
## YPEL1 0.000000e+00 4.027619e+01 0.000000e+00 0.000000e+00
## YWHAE 7.966660e+01 7.356225e+01 9.998782e+01 1.058337e+02
## YWHAH 1.611435e+02 8.127311e+01 1.630869e+02 1.706133e+03
## ZDHHC1 3.167680e+01 0.000000e+00 0.000000e+00 0.000000e+00
## ZFAND6 5.709186e+01 8.866025e+01 3.542941e+01 7.574516e+01
## ZFP36 1.336661e+03 1.144461e+03 1.623968e+03 1.121272e+02
## ZFP36L1 2.627076e+01 4.477157e+01 7.471008e+01 7.678143e+01
## ZGLP1 7.527045e+00 0.000000e+00 1.405676e+01 2.032610e+02
## ZMAT5 3.391060e+01 3.598812e+01 2.742478e+01 0.000000e+00
## ZMYND8 2.326553e+00 2.627470e+00 1.376282e+01 0.000000e+00
## ZNF106 1.129188e+01 8.343958e+00 9.481860e+00 5.125814e+00
## ZNF689 4.335204e+00 0.000000e+00 1.500716e+01 0.000000e+00
## ZNF778 9.686924e-01 0.000000e+00 0.000000e+00 9.353821e+00
## ZNHIT3 6.621318e+01 2.827091e+01 2.602687e+01 0.000000e+00Ciber_SE <- deconv_TME(MEL_GSE91061,
method = "CIBERSORT",
sig_matrix=ref_sig_mtr)
diff_TME(Ciber_SE,feature = rownames(Ciber_SE)[1:8])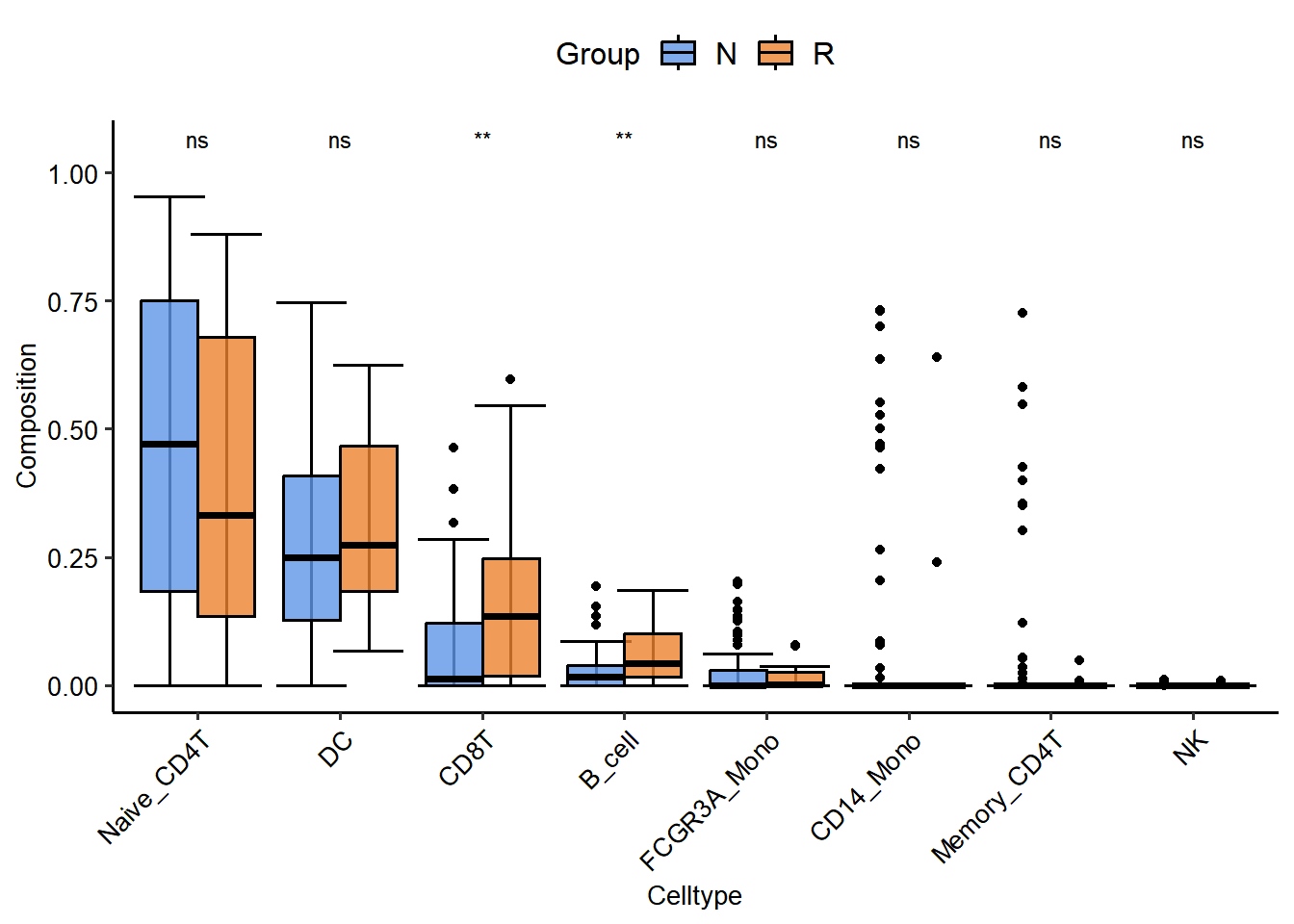
📝 More details about TME analysis
TIMER is a comprehensive tool for systematical analysis of immune infiltrates across diverse cancer types.
CIBERSORT is an analytical tool from the Alizadeh Lab and Newman Lab to impute gene expression profiles and provide an estimation of the abundances of member cell types in a mixed cell population, using gene expression data.
MCPCounter is called the Microenvironment Cell Populations-counter. It allows the robust quantification of the absolute abundance of eight immune and two stromal cell populations in heterogeneous tissues from transcriptomic data.
xCell is a gene signature-based method learned from thousands of pure cell types from various sources. xCell applies a novel technique for reducing associations between closely related cell types.
IPS uses an analytical strategy to provide comprehensive view of 28 TIL subpopulations including effector and memory T cells and immunosuppressive cells (Tregs, MDSCs).
EPIC is called Estimating the Proportion of Immune and Cancer cells. It compares the level of expression of genes in a tumor with a library of the gene expression profiles from specific cell types that can be found in tumors and uses this information to predict the abundance of each cell type.
ESTIMATE is described as ‘Estimation of STromal and Immune cells in MAlignant Tumours using Expression data’. It is a method that utilizes gene expression signatures to infer the proportion of stromal and immune cells within tumor samples.
ABIS is called ABsolute Immune Signal (ABIS) deconvolution. ABIS performs absolute deconvolution on RNA-Seq and microarray data.
ConsensusTME uses a consensus approch to generate cancer-specific signatres for TME cell types, and utilizes ssGSEA framework to estimate the relative abundance of these cell types.
quanTIseq is a computational pipeline for the quantification of the Tumor Immune contexture from human RNA-seq data.