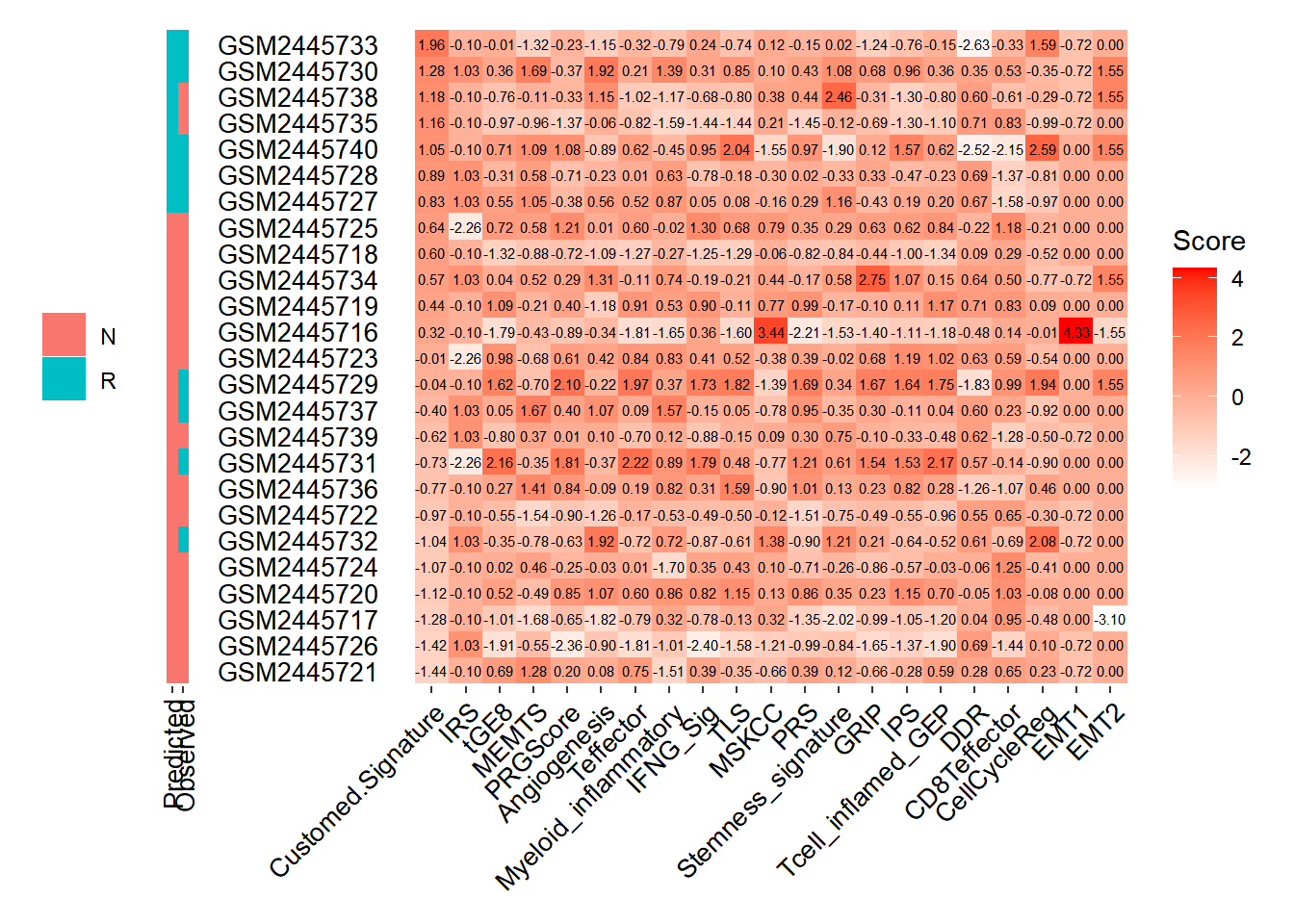
Chapter 6 🔮 Response Prediction
pred_response function predicts the immunotherapy response for the patients from gene expression data using our pre-trained machine learning models or public gene expression signatures.
The ipt signature is derived from multi-modal data, which includes bulk RNA-seq, tumor microenvironment information, and clinical data, collected from 275 melanoma patients who underwent Immune checkpoint blockade treatment. A Random Forest model was constructed to conduct feature selection and create the signature.
pred_response(SE=MEL_GSE93157,Signature = ipt,
method = "Weighted_mean",threshold = 0.8,
PT_drop = FALSE,sort_by = "Customed.Signature",
group_by = "Customed.Signature",show.Observed = TRUE,
rankscore = FALSE)## 2 Signature genes are not found in expression matrix. The function can execute properly, but the performance of the model may be compromised.
## 2 Signature genes are not found in expression matrix. The function can execute properly, but the performance of the model may be compromised.## 1 Signature genes are not found in expression matrix. The function can execute properly, but the performance of the model may be compromised.## 3 Signature genes are not found in expression matrix. The function can execute properly, but the performance of the model may be compromised.## 1 Signature genes are not found in expression matrix. The function can execute properly, but the performance of the model may be compromised.## B.cell T.cell.CD4. T.cell.CD8. Neutrophil Macrophage Myeloid.dentritic.cell IRS tGE8 MEMTS PRGScore Angiogenesis Teffector Myeloid_inflammatory IFNG_Sig TLS MSKCC LMRGPI PRS Stemnesssignatures Riskscore IPS Tcell_inflamed_GEP DDR CD8Teffector CellCycleReg PanFTBRs EMT1 EMT2 EMT3 does not exist in expression matrix.