The KEGG FTP service has not been freely available for academic use since 2012, and there are many software packages using outdated KEGG annotation data. The clusterProfiler package supports downloading the latest online version of KEGG data using the KEGG website , which is freely available for academic users. Both KEGG pathways and modules are supported in clusterProfiler .
Supported organisms
The clusterProfiler package supports all organisms that have KEGG annotation data available in the KEGG database. Users should pass an abbreviation of academic name to the organism parameter. The full list of KEGG supported organisms can be accessed via http://www.genome.jp/kegg/catalog/org_list.html . KEGG Orthology (KO) Database is also supported by specifying organism = "ko".
The clusterProfiler package provides the search_kegg_organism() function to help search for supported organisms.
library (clusterProfiler)search_kegg_organism ('ece' , by= 'kegg_code' )
kegg_code scientific_name common_name
1294 ece Escherichia coli O157:H7 EDL933 EHEC
8299 ecec Enterococcus cecorum Enterococcus cecorum
<- search_kegg_organism ('Escherichia coli' , by= 'scientific_name' )dim (ecoli)
kegg_code scientific_name common_name
1288 eco Escherichia coli K-12 MG1655 Escherichia coli K-12 MG1655
1289 ecj Escherichia coli K-12 W3110 Escherichia coli K-12 W3110
1290 ecd Escherichia coli K-12 DH10B Escherichia coli K-12 DH10B
1291 ebw Escherichia coli K-12 BW2952 Escherichia coli K-12 BW2952
1292 ecok Escherichia coli K-12 MDS42 Escherichia coli K-12 MDS42
1293 ecoc Escherichia coli K-12 C3026 Escherichia coli K-12 C3026
KEGG pathway over-representation analysis
data (geneList, package= "DOSE" )<- names (geneList)[abs (geneList) > 2 ]<- enrichKEGG (gene = gene,organism = 'hsa' ,pvalueCutoff = 0.05 )head (kk)
category
hsa04110 Cellular Processes
hsa04114 Cellular Processes
hsa04218 Cellular Processes
hsa04061 Environmental Information Processing
hsa03320 Organismal Systems
hsa04814 Cellular Processes
subcategory ID
hsa04110 Cell growth and death hsa04110
hsa04114 Cell growth and death hsa04114
hsa04218 Cell growth and death hsa04218
hsa04061 Signaling molecules and interaction hsa04061
hsa03320 Endocrine system hsa03320
hsa04814 Cell motility hsa04814
Description
hsa04110 Cell cycle
hsa04114 Oocyte meiosis
hsa04218 Cellular senescence
hsa04061 Viral protein interaction with cytokine and cytokine receptor
hsa03320 PPAR signaling pathway
hsa04814 Motor proteins
GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
hsa04110 15/109 158/9561 0.09493671 8.327430 9.973035 2.555067e-10
hsa04114 10/109 139/9561 0.07194245 6.310475 6.772508 3.821301e-06
hsa04218 10/109 157/9561 0.06369427 5.586981 6.223014 1.131941e-05
hsa04061 8/109 100/9561 0.08000000 7.017248 6.495457 1.719645e-05
hsa03320 7/109 76/9561 0.09210526 8.079068 6.653414 2.360914e-05
hsa04814 10/109 197/9561 0.05076142 4.452568 5.258054 7.963848e-05
p.adjust qvalue
hsa04110 8.917183e-08 8.917183e-08
hsa04114 6.668170e-04 6.668170e-04
hsa04218 1.316825e-03 1.316825e-03
hsa04061 1.500390e-03 1.500390e-03
hsa03320 1.647918e-03 1.647918e-03
hsa04814 4.632305e-03 4.632305e-03
geneID
hsa04110 9133/10403/891/81620/4085/890/9319/1111/4174/983/8318/7272/9212/9232/991
hsa04114 9133/891/4085/3708/6790/983/51806/5241/9232/991
hsa04218 9133/4605/776/2305/891/890/3708/1111/983/51806
hsa04061 6373/6362/10563/1524/4283/6355/3627/9547
hsa03320 5105/9370/9415/3158/5346/4312/2167
hsa04814 81930/146909/10112/9493/4629/3832/7802/24137/3833/1062
Count
hsa04110 15
hsa04114 10
hsa04218 10
hsa04061 8
hsa03320 7
hsa04814 10
Input ID type can be kegg, ncbi-geneid, ncbi-proteinid or uniprot (see also Section 17.1.2 enrichGO(), there is no readable parameter for enrichKEGG(). However, users can use the setReadable() function, see also Section 17.2 OrgDb available for the species.
KEGG pathway gene set enrichment analysis
<- gseKEGG (geneList = geneList,organism = 'hsa' ,minGSSize = 120 ,pvalueCutoff = 0.05 ,verbose = FALSE )head (kk2)
ID Description setSize
hsa04110 hsa04110 Cell cycle 139
hsa05169 hsa05169 Epstein-Barr virus infection 195
hsa04613 hsa04613 Neutrophil extracellular trap formation 130
hsa05166 hsa05166 Human T-cell leukemia virus 1 infection 203
hsa04218 hsa04218 Cellular senescence 141
hsa04114 hsa04114 Oocyte meiosis 96
enrichmentScore NES pvalue p.adjust qvalue rank
hsa04110 0.6637551 2.818492 3.476518e-12 3.754640e-10 3.754640e-10 1155
hsa05169 0.4335116 1.932132 2.213318e-07 1.195192e-05 1.195192e-05 2820
hsa04613 0.4496570 1.927342 6.963115e-06 2.506721e-04 2.506721e-04 2575
hsa05166 0.3882568 1.762110 1.601941e-05 4.325240e-04 4.325240e-04 1955
hsa04218 0.4115945 1.743754 8.316752e-05 1.143351e-03 1.143351e-03 1155
hsa04114 0.4396853 1.737405 2.341679e-04 2.107511e-03 2.107511e-03 896
leading_edge
hsa04110 tags=36%, list=9%, signal=33%
hsa05169 tags=39%, list=23%, signal=31%
hsa04613 tags=37%, list=21%, signal=30%
hsa05166 tags=26%, list=16%, signal=22%
hsa04218 tags=17%, list=9%, signal=16%
hsa04114 tags=20%, list=7%, signal=19%
core_enrichment
hsa04110 8318/991/9133/10403/890/983/4085/81620/7272/9212/1111/9319/891/4174/9232/4171/993/990/5347/701/9700/898/23594/4998/9134/4175/4173/10926/6502/994/699/4609/5111/26271/1869/1029/8317/4176/2810/3066/1871/1031/9088/995/1019/4172/5885/11200/7027/1875
hsa05169 3627/890/6890/9636/898/9134/6502/6772/3126/3112/4609/917/5709/1869/3654/919/915/4067/4938/864/4940/5713/5336/11047/3066/54205/1871/578/1019/637/916/3383/4939/10213/23586/4793/5603/7979/7128/6891/930/5714/3452/6850/5702/4794/7124/3569/7097/5708/2208/8772/3119/5704/7186/5971/3135/1380/958/5610/55080/4792/10018/8819/3134/10379/9641/1147/5718/6300/3109/811/5606/2923/3108/5707/1432
hsa04613 820/366/51311/64581/3015/85236/55506/8970/8357/1535/2359/5336/4688/92815/3066/8336/292/1991/3689/8345/5603/4689/5880/10105/1184/6404/3018/6850/5604/3014/7097/1378/8290/1536/834/5605/1183/728/2215/8335/5594/9734/3674/5578/5582/7417/8331/6300
hsa05166 991/9133/890/4085/7850/1111/9232/8061/701/9700/898/4316/9134/3932/3559/3126/3112/4609/3561/917/1869/1029/915/114/2005/5902/55697/1871/1031/2224/292/1019/3689/916/3383/11200/706/3600/6513/3601/468/5604/7124/1030/3569/4049/4055/10393/3119/5901/5971/1959/3135
hsa04218 2305/4605/9133/890/983/51806/1111/891/993/3576/1978/898/9134/4609/1869/1029/22808/1871/5499/91860/292/1019/11200/1875
hsa04114 991/9133/983/4085/51806/6790/891/9232/5347/9700/898/9134/699/26271/114/5499/91860/9088/995
KEGG module over-representation analysis
KEGG Module is a collection of manually defined functional units. In some situations, KEGG Modules have a more straightforward interpretation.
<- enrichMKEGG (gene = gene,organism = 'hsa' ,pvalueCutoff = 1 ,qvalueCutoff = 1 )head (mkk)
ID
M00912 M00912
M00095 M00095
M00053 M00053
M00938 M00938
M00003 M00003
M00049 M00049
Description
M00912 NAD biosynthesis, tryptophan => quinolinate => NAD
M00095 C5 isoprenoid biosynthesis, mevalonate pathway
M00053 Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP
M00938 Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP
M00003 Gluconeogenesis, oxaloacetate => fructose-6P
M00049 Adenine ribonucleotide biosynthesis, IMP => ADP,ATP
GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
M00912 2/9 12/833 0.16666667 15.425926 5.257534 0.006480816
M00095 1/9 10/833 0.10000000 9.255556 2.743252 0.103471964
M00053 1/9 11/833 0.09090909 8.414141 2.585477 0.113276038
M00938 1/9 14/833 0.07142857 6.611111 2.211517 0.142119005
M00003 1/9 18/833 0.05555556 5.141975 1.855600 0.179278025
M00049 1/9 19/833 0.05263158 4.871345 1.782977 0.188341212
p.adjust qvalue geneID Count
M00912 0.2851559 0.2851559 23475/3620 2
M00095 1.0000000 1.0000000 3158 1
M00053 1.0000000 1.0000000 6241 1
M00938 1.0000000 1.0000000 6241 1
M00003 1.0000000 1.0000000 5105 1
M00049 1.0000000 1.0000000 26289 1
KEGG module gene set enrichment analysis
<- gseMKEGG (geneList = geneList,organism = 'hsa' ,pvalueCutoff = 1 )head (mkk2)
ID Description setSize
M00912 M00912 NAD biosynthesis, tryptophan => quinolinate => NAD 8
M00001 M00001 Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate 24
M00004 M00004 Pentose phosphate pathway (Pentose phosphate cycle) 9
M00002 M00002 Glycolysis, core module involving three-carbon compounds 11
M00099 M00099 Sphingosine biosynthesis 9
M00938 M00938 Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP 10
enrichmentScore NES pvalue p.adjust qvalue rank
M00912 0.8509100 1.908409 0.000800003 0.03520013 0.01946038 679
M00001 0.5739035 1.761196 0.005706550 0.12554409 0.06940701 2886
M00004 0.7450305 1.690370 0.010384991 0.15231320 0.08420631 1750
M00002 0.6421781 1.631769 0.045055261 0.33040525 0.18266444 1381
M00099 -0.7084736 -1.602108 0.020187880 0.22206668 0.12276950 2125
M00938 0.6648002 1.583961 0.037161361 0.32701998 0.18079290 648
leading_edge
M00912 tags=62%, list=5%, signal=59%
M00001 tags=54%, list=23%, signal=42%
M00004 tags=67%, list=14%, signal=57%
M00002 tags=55%, list=11%, signal=49%
M00099 tags=56%, list=17%, signal=46%
M00938 tags=40%, list=5%, signal=38%
core_enrichment
M00912 23475/3620/6999/8564/8942
M00001 5214/3101/2821/7167/2597/5230/2023/5223/5315/3099/5232/2027/5211
M00004 2821/5226/7086/2539/6888/22934
M00002 7167/2597/5230/2023/5223/5315
M00099 253782/29956/427/55304/79603
M00938 6241/7298/4830/1841
Visualize enriched KEGG pathways
The enrichplot package implements several methods to visualize enriched terms. Most of them are general methods that can be used on GO, KEGG, MSigDb, and other gene set annotations. Here, we introduce the clusterProfiler::browseKEGG() and pathview::pathview() functions to help users explore enriched KEGG pathways with genes of interest.
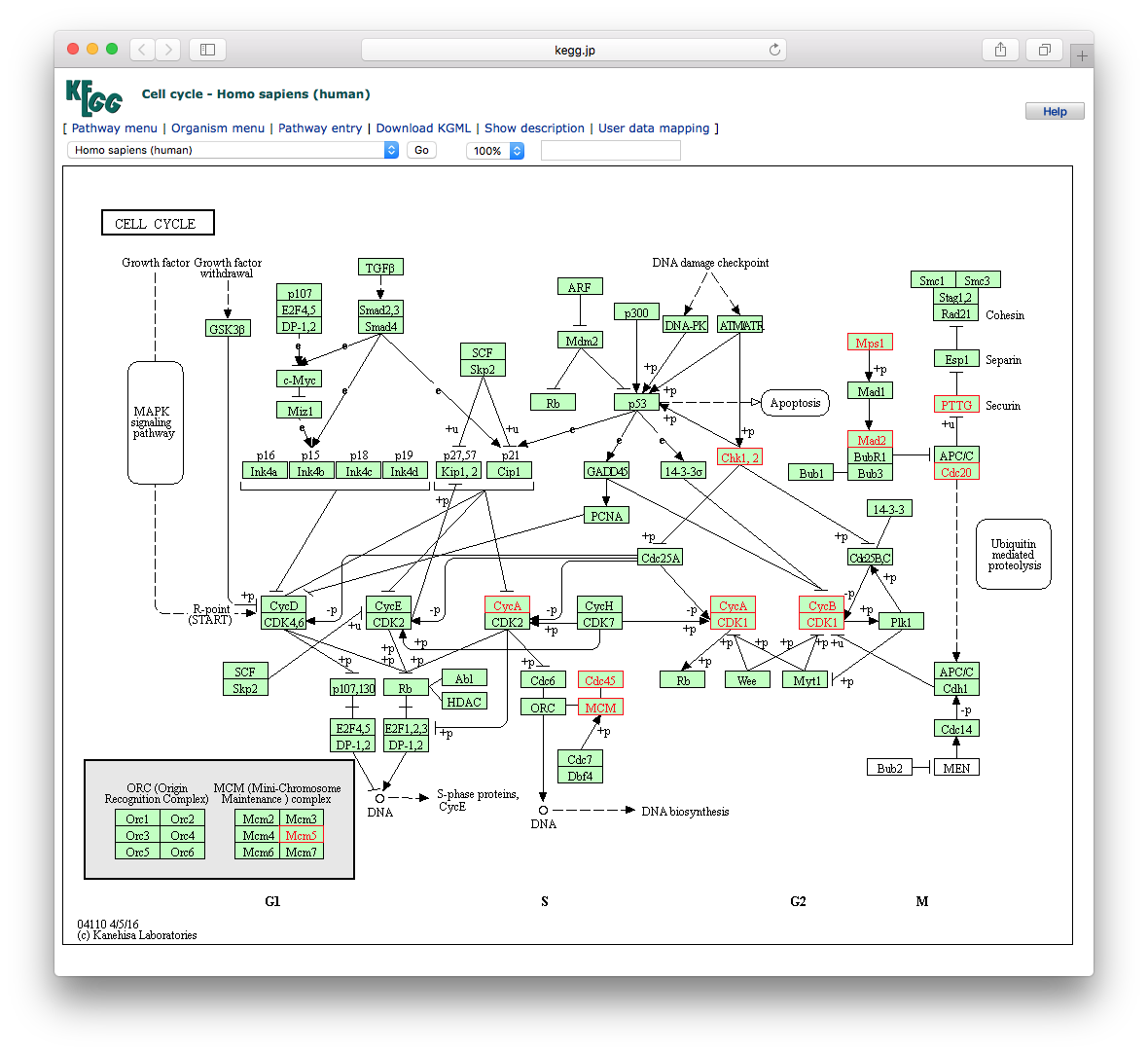
To view the KEGG pathway, users can use the browseKEGG() function, which will open a web browser and highlight enriched genes.
browseKEGG (kk, 'hsa04110' )
(ref:browseKEGGscap) Explore selected KEGG pathway.
(ref:browseKEGGcap) Explore selected KEGG pathway. Differentially expressed genes that are enriched in the selected pathway will be highlighted.
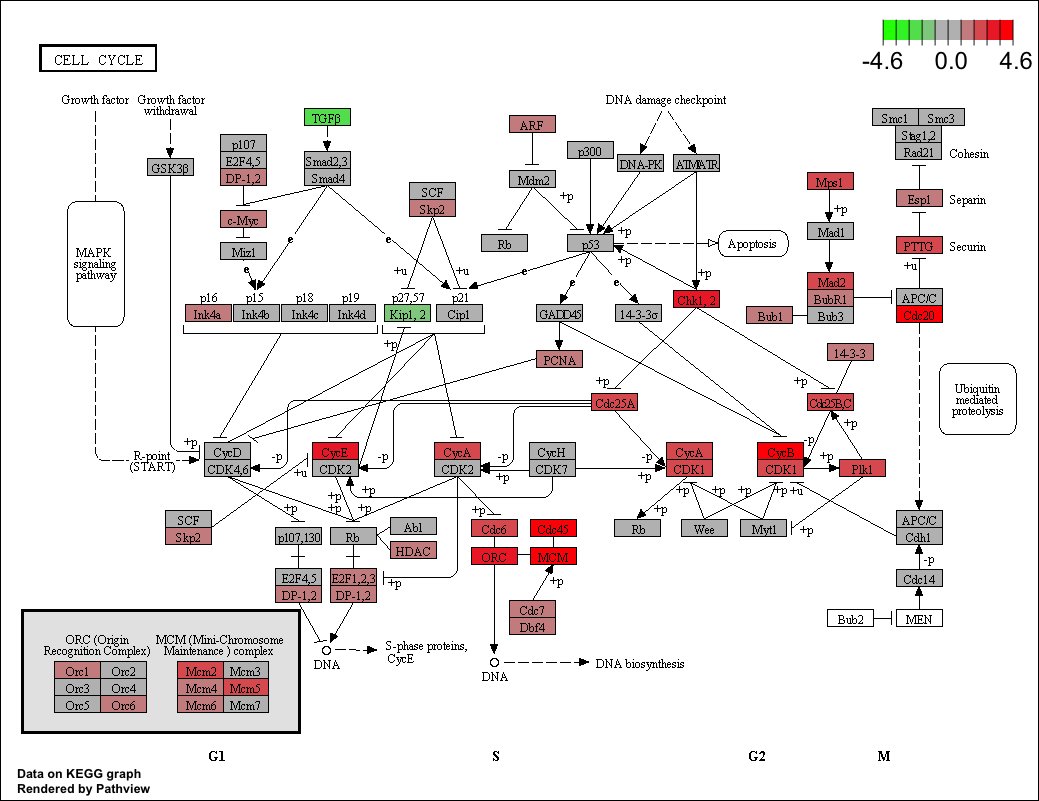
Users can also use the pathview() function from the pathview (Luo and Brouwer 2013 ) to visualize enriched KEGG pathways identified by the clusterProfiler package (Yu et al. 2012 ) .
The following example illustrates how to visualize the “hsa04110” pathway, which was enriched in our previous analysis.
library ("pathview" )<- pathview (gene.data = geneList,pathway.id = "hsa04110" ,species = "hsa" ,limit = list (gene= max (abs (geneList)), cpd= 1 ))
(ref:pathviewscap) Visualize selected KEGG pathway by pathview().
(ref:pathviewcap) Visualize selected KEGG pathway by pathview(). Gene expression values can be mapped to gradient color scale.
References
Luo, Weijun, and Cory Brouwer. 2013.
“Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization.” Bioinformatics 29 (July): 1830–31.
https://doi.org/10.1093/bioinformatics/btt285 .
Yu, Guangchuang, Le-Gen Wang, Yanyan Han, and Qing-Yu He. 2012.
“clusterProfiler: An r Package for Comparing Biological Themes Among Gene Clusters.” OMICS: A Journal of Integrative Biology 16 (5): 284–87.
https://doi.org/10.1089/omi.2011.0118 .