📖 Introduction
hello
🎯 Motivation
The book is meant as a guide for mining biological knowledge to elucidate or interpret molecular mechanisms using a suite of R packages, including ChIPseeker, clusterProfiler, DOSE, enrichplot, GOSemSim, meshes and ReactomePA. Hence, if you are starting to read this book, we assume you have a working knowledge of how to use R.
📝 Citation
If you use the software suite in published research, please cite the most appropriate paper(s) from this list:
- T Wu#, E Hu#, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo*, G Yu*. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021, 2(3):100141. doi: 10.1016/j.xinn.2021.100141
- G Yu*. Gene Ontology Semantic Similarity Analysis Using GOSemSim. In: Kidder B. (eds) Stem Cell Transcriptional Networks. Methods in Molecular Biology. 2020, 2117:207-215. Humana, New York, NY. doi: 10.1007/978-1-0716-0301-7_11
- G Yu*. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics. 2018, 34(21):3766–3767. doi: 10.1093/bioinformatics/bty410
- G Yu, QY He*. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Molecular BioSystems. 2016, 12(2):477-479. doi: 10.1039/C5MB00663E
- G Yu*, LG Wang, and QY He*. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015, 31(14):2382-2383. doi: 10.1093/bioinformatics/btv145
- G Yu*, LG Wang, GR Yan, QY He*. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics. 2015, 31(4):608-609. doi: 10.1093/bioinformatics/btu684
- G Yu, LG Wang, Y Han and QY He*. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287. doi: 10.1089/omi.2011.0118
- G Yu, F Li, Y Qin, X Bo*, Y Wu, S Wang*. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010, 26(7):976-978. doi: 10.1093/bioinformatics/btq064
📚 Book structure
- Part 1 (Semantic similarity analysis) describes GOSemSim, DOSE and meshes packages for measuring semantic similarity of genes or gene products based on Gene Ontology, Disease Ontology and Medical Subject Headings.
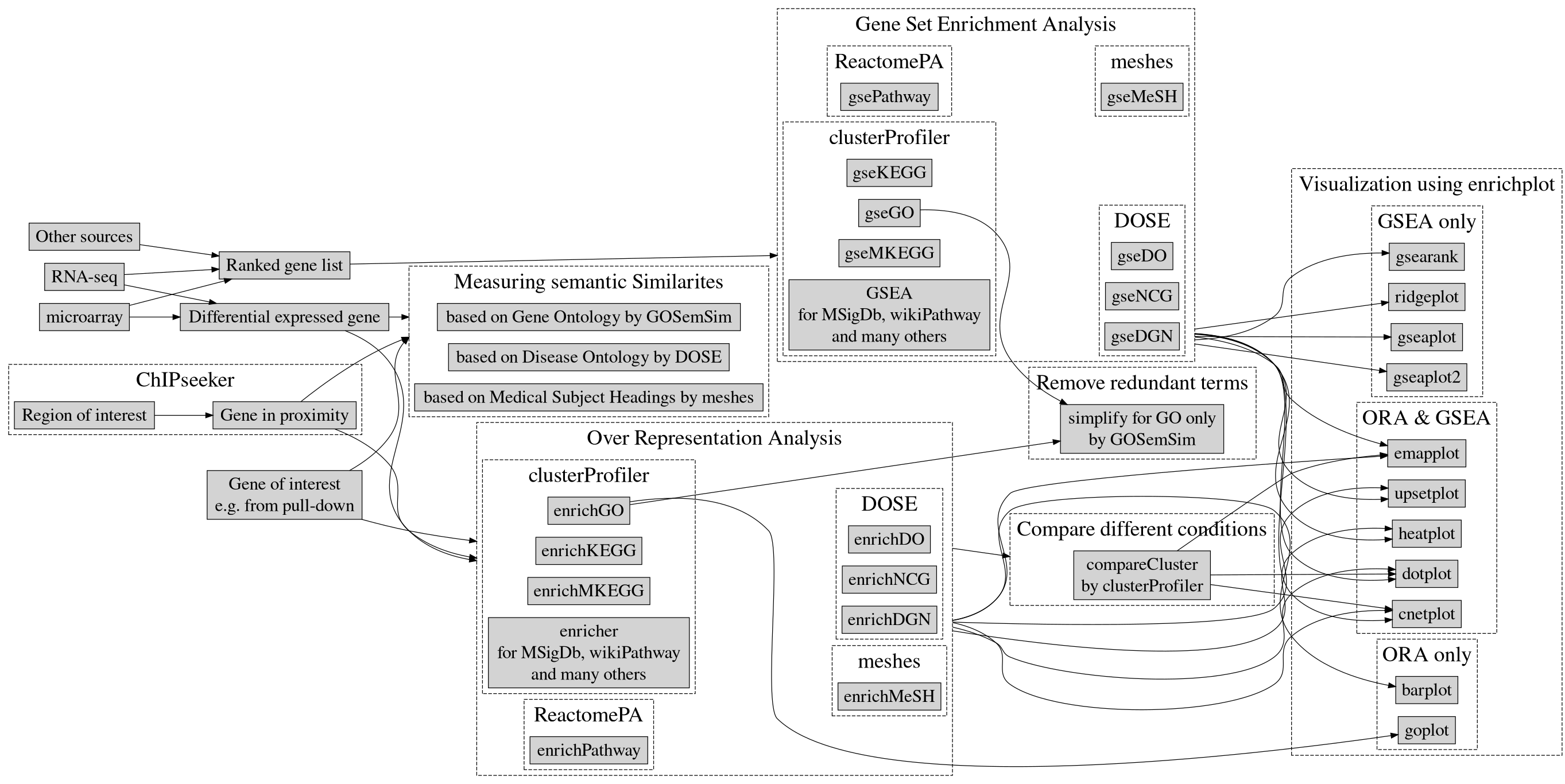
- Part 2 (Enrichment analysis) introduces over-reprensentation analysis and gene set enrichment analysis using clusterProfiler (supports GO, KEGG, MSigDb, WikiPathway, and many others via universal interface), DOSE (DO, Disease-Gene Network, Network of Cancer Genes), meshes (MeSH), and ReactomePA (Reactome pathway). Functional enrichment analysis of Genomic coordination is supported via ChIPseeker and comparison among multiple conditions is also supported by clusterProfiler. We implemented a number of visualization methods in the enrichplot package to help users to interpret their results.
- Part 3 (Miscellaneous topics) describes useful utilities including translating gene IDs and manipulating enrichment results.
💖 Want to help?
The book’s source code is hosted on GitHub, at https://github.com/YuLab-SMU/biomedical-knowledge-mining-book. Any feedback on the book is very welcome. Feel free to open an issue on GitHub or send me a pull request if you notice typos or other issues (I’m not a native English speaker ;) ).






