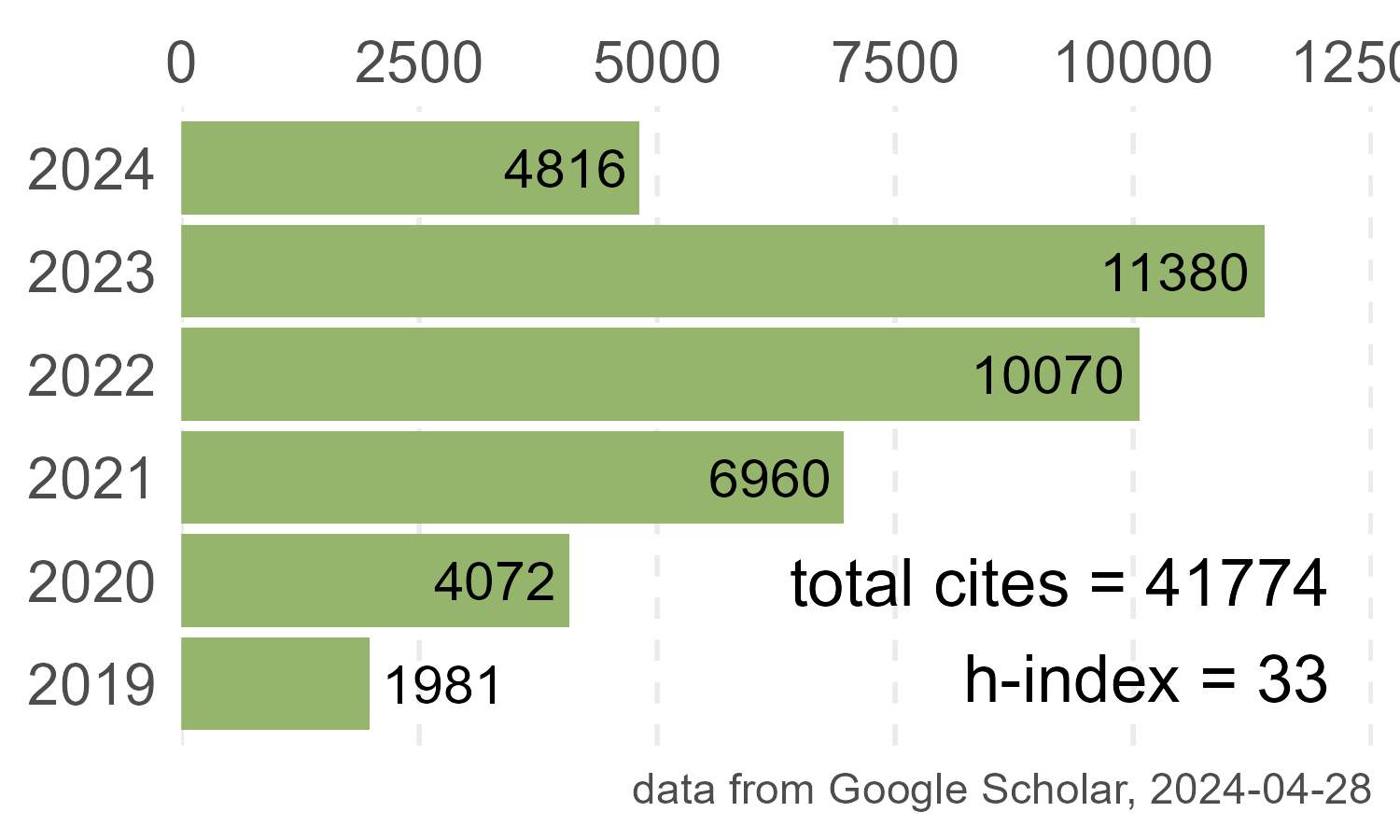
Publications
|
|
2026 Link to heading
- Y Feng#, L Zheng#, W Tang#, P Wang, Y Lai, J Qin, C Zhou, X Zhang, M Yang, L Jie*, G Yu*, H Ren*, Q Huang*. Gut metabolite indole-3-acetic acid aggravates neuropsychiatric lupus via the AHR/STAT3 pathway in microglia. Communications Biology. 2026, 9(1):281.
- B Liu, Y Liu, S Xu, Q Wu, D Wu, Y Liao, Y Mai, M Zheng, S Wang, L Zhan, Y Chen, Z Huang, X Luo, Z Xie, X Pan, G Yu*, L Xie*. EasyMultiProfiler: An Efficient Multi-Omics Data Integration and Analysis Workflow for Microbiome Research. SCIENCE CHINA Life Sciences. 2026, 69(2):321-330. (封面文章, Cover Article)
- Q Wang#, L Deng#, S Xu#, P Guo, H Zhu, H Ge, Y Gong, G Du, K Huang, C Su, R Wang, Y Qiu*, G Yu*. Comparison of Illumina NovaSeq 6000 and GeneMind SURFSeq 5000 platforms for single‐cell spatial transcriptomics of mouse brain and lung. Interdisciplinary Medicine. 2026, 4(1):e70067.
- G Yu*. Background bias in functional enrichment analysis: Insights from clusterProfiler. The Innovation Life. 2026, 4(1):100181. (封面导读:Insights form clusterProfiler)
2025 Link to heading
- Y Peng, S Jiang, Y Song, P Luo, J Li, D Hu, JG Zhou, G Yu*, T Xu*, S Wang*. ggalign: Bridging the Grammar of Graphics and Biological Multilayered Complexity. Advanced Science. 2025, 12(44): e07799.
- S Wen#, X Du#, M Zhu#, C Huang, Z Lin, M Li, B Hu, X Wang, F Jiang, G Zhou, W Zhu, G Yu*, C Xu*, B Shen*. ALCAM-CD6 axis suppression: a key determinant of immune-mediated metastasis recurrence in stage III non-small cell lung cancer. Journal for ImmunoTherapy of Cancer. 2025, 13(10):e010416.
- P Li#, L Xie#, H Zheng#, Y Feng#, F Mai, W Tang, J Wang, Z Lan, S Lv, T Jayawardana, S Koentgen, S Xu, Z Wan, Y Chen, H Xu, S Shen, F Zhang, Y Yang, G Hold, F He*, E M. El-Omar*, G Yu*, X Chen*. Gut microbial-derived 3,4-dihydroxyphenylacetic acid ameliorates reproductive phenotype of polycystic ovary syndrome. iMeta. 2025, 4(5):e70065.
- X Lu, K Li, Z Li, A Lin, L Zhao, R Shen, Z Xu, J Gao, D Lv, Y Zhang, T Ye, J Shen, Y Chen, H Huang, Z Hao, D Zeng, H Wang, S Guo, W Wang, Y Xiong, Y Li, H Li, J Gao, Q Liu, B Wei, J Shi, S Cheng, G Li, Y Tang, E Lin, H Xu, G Feng, Y He, Y Sun, X Liu, Y Wang, W Song, J Song, S Tian, Y Zhang, J Zhang, Z Xu, C Song, Y Zhang, H Wu, C Gao, E Hu, C Yang, J Lou, D Wang, X Wang, P Luo*, G Yu*, Y Ge*. FigureYa: A Standardized Visualization Framework for Enhancing Biomedical Data Interpretation and Research Efficiency. iMetaMed. 2025, 1(1):e70005. Cover story
- S Xu#, Q Wang#, S Wen#, J Li#, N He#, M Li, T Hackl, R Wang, D Zeng, S Wang, S Li, CH Gao, L Zhou, S Tao, Z Xie, L Deng, G Yu*. aplot: Simplifying the creation of complex graphs to visualize associations across diverse data types. The Innovation. 2025, 6(9):100958. (封面导读:Complex composite graphics)
- Y Gao#, B Li#, Y Jin#, J Cheng#, W Tian, L Ying, L Hong, S Xin, B Lin, C Liu, X Sun, J Zhang, H Zhang, J Xie, X Deng, X Dai, L Liu, Y Zheng, P Zhao, G Yu*, W Fang*, X Bao*. Spatial multi-omics profiling of breast cancer oligo-recurrent lung metastasis. Oncogene. 2025, 44(27):2268-2282.
- Q Wang, H Zhu, L Deng, S Xu, W Xie, M Li, R Wang, L Tie, L Zhan, G Yu*. Spatial Transcriptomics: Biotechnologies, Computational Tools, and Neuroscience Applications. Small Methods. 2025, 9(5):2401107. 卷首插图文章Frontispiece
- Y Tu#*, S Xu#, H Shu, X Wang, W Li, G Yu*, Li He*. 16S rRNA Sequencing Reveals Dysbiosis of Skin Microbiome Associated With Disease Severity in Chronic Actinic Dermatitis. Photodermatology, Photoimmunology & Photomedicine. 2025, 41(3):e70022.
- M Chen#, X Luo#, S Xu#, L Li, J Li, Z Xie, Q Wang, Y Liao, B Liu, W Liang, K Mo, Q Song, X Chen*, TTY Lam*, G Yu*. Scalable method for exploring phylogenetic placement uncertainty with custom visualizations using treeio and ggtree. iMeta. 2025, 4(1):e269.
2024 Link to heading
- D Zeng#, Y Fang#, W Qiu; P Luo, S Wang; R Shen, W Gu, X Huang, Q Mao; G Wang, Y Lai; X Xu, M Shi, Z Wu*, G Yu*, W Liao*. Enhancing Immuno-Oncology Investigations Through Multidimensional Decoding of Tumour Microenvironment with IOBR 2.0. Cell Reports Methods. 2024, 4(12):100910. (IF = 4.5,二区)
- M Luo*#, W Yang#, L Bai#, L Zhang#, JW Huang#, Y Cao#, Y Xie#, L Tong, H Zhang, L Yu, LW Zhou, Y Shi, P Yu, Z Wang, Z Yuan, P Zhang, Y Zhang, F Ju, H Zhang, F Wang, Y Cui, J Zhang, G Jia, D Wan, C Ruan, Y Zeng, P Wu, Z Gao, W Zhao, Y Xu, G Yu, C Tian, LN Jin, J Dai*, B Xia*, B Sun*, F Chen*, YZ Gao*, H Wang*, B Wang*, D Zhang*, X Cao*, H Wang*, T Huang*. Artificial intelligence for life sciences: A comprehensive guide and future trends. The Innovation Life. 2024, 2(4):100105.
- Z Miao#, T Tian#, W Chen#, Q Wang#, L Ma#, D Zhang#, M Xie#, Z Yu#, X Guo, G Bai, S Zhao, X Chen, W Wang, Y Gao, S Guo*, M Luo*, L Yuan*, C Tian*, L Wu*, G Yu*, D Zhang*, S Sun*. Spatial Resolved Transcriptomics: Computational Insights into Gene Transcription Across Tissue and Organ Architecture in Diverse Applications. The Innovation Life. 2024, 2(4):100097.
- G Yu*. Thirteen years of clusterProfiler. The Innovation. 2024, 5(6):100722. (IF = 25.7,一区)
- S Xu#, E Hu#, Y Cai#, Z Xie#, X Luo#, L Zhan, W Tang, Q Wang, B Liu, R Wang, W Xie, T Wu, L Xie, G Yu*. Using clusterProfiler to characterise Multi-Omics Data. Nature Protocols. 2024, 19(11):3292-3320. (IF = 16.0,一区)
- S Wang, G Yu, L Xie*. Asymmetric Cell Division and Satellite Cell Fate Regulation in Skeletal Muscle Aging and Disease. Health and Metabolism. 2024, 1(1):5.
- Y Chen#, LN He#, Y Zhang, J Gong, S Xu, Y Shu, D Zhang*, G Yu*, Z Zuo*. tigeR: Tumor Immunotherapy Gene Expression Data Analysis R package. iMeta. 2024, 3(5):e229. (IF = 33.2,一区)
- D Wang, G Chen, L Li, S Wen, Z Xie, X Luo, L Zhan, S Xu, J Li, R Wang, Q Wang*, G Yu*. Reducing language barriers, promoting information absorption, and communication using fanyi. Chinese Medical Journal. 2024, 137(16):1950-1956. (IF = 7.3,二区)
- XK Ma#, Y Yu#, T Huang#*, D Zhang, C Tian, W Tang, M Luo, P Du*, G Yu*, L Yang*. Bioinformatics Software Development: principles and future directions. The Innovation Life. 2024, 2(3):100083.
- Y Yang#, S Xu#, Y Hong, Y Cai, W Tang, J Wang, B Shen*, H Zong*, G Yu*. Computational modeling for medical data: from data collection to knowledge discovery. The Innovation Life. 2024, 2(3):100079.
- L Zhan#, X Luo#, W Xie#, XA Zhu#, Z Xie, J Lin, L Li, W Tang, R Wang, L Deng, Y Liao, B Liu, Y Cai, Q Wang, S Xu*, G Yu*. shinyTempSignal: an R shiny application for exploring temporal and other phylogenetic signals. Journal of Genetics and Genomics. 2024, 51(7):762-768. (IF = 7.1,一区)
- L Li, W Xie, L Zhan, S Wen, X Luo, S Xu, Y Cai, W Tang, Q Wang, M Li, Z Xie, L Deng, H Zhu, G Yu*. Resolving Tumor Evolution: A Phylogenetic Approach. Journal of the National Cancer Center. 2024, 4(2):97-106. (IF = 9.4,三区)
- Q Wang, W Tang, L Chen, L Deng, G Yu*. Decoding the Brain: Unveiling Comprehensive Cellular Atlases through Multiomics Mega Data. The Innovation. 2024, 5(4):100637. (IF = 25.7,一区)
- Y Cai#, W Hu#, Y Pei, H Zhao*, G Yu*. Encoding biological metaverse: Advancements and challenges in neural fields from macroscopic to microscopic. The Innovation. 2024, 5(3):100627. (IF = 25.7,一区)
- Y Lai#, W Tang#, X Luo, H Zheng, Y Zhang*, M Wang*, G Yu*, Min Yang*. Gut microbiome and metabolome to discover pathogenic bacteria and probiotics in ankylosing spondylitis. Frontiers in Immunology. 2024, 15:1369116. (IF = 5.9,二区)
- CH Gao, C Chen, T Akyol, A Dusa, G Yu, B Cao, P Cai*. ggVennDiagram: intuitive Venn diagram software extended. iMeta. 2024, 3(1):e177. (IF = 33.2,一区)
- Y Ye#, MH Shum#, JL Tsui#, G Yu#, DK Smith, H Zhu, JT Wu, Y Guan, TTY Lam*. Robust expansion of phylogeny for fast-growing genome sequence data. PLoS Computational Biology. 2024, 20(2):e1011871. (IF = 3.6,二区)
- S Wen, S Mo, J Zhou, Y Lv, K Khazaie*, G Yu*. Single-cell and spatial-omics in delineating immune-related diseases. Frontiers in Cell and Developmental Biology. 2024, 12:1365242. (IF = 4.3,二区)
2023 Link to heading
- X Zhang, B Liang, Y Huang, H Meng, Z Li, J Du, L Zhou, Y Zhong, B Wang, X Lin, G Yu, X Chen, W Lu, ZS Chen, X Yang, Z Huang*. Behind the Indolent Facade: Uncovering the Molecular Features and Malignancy Potential in Lung Minimally Invasive Adenocarcinoma by Single-Cell Transcriptomics. Advanced Science. 2023, 10(36):2303753. (IF = 14.3,一区)
- M Kang, F Lin, Z Jiang, X Tan, X Lin, Z Liang, C Xiao, Y Xia, W Guan, Z Yang, G Yu, M Zanin, S Tang*, SS Wong*. The impact of pre-existing influenza antibodies and inflammatory status on the influenza vaccine responses in older adults. Influenza and Other Respiratory Viruses. 2023, 17(7):e13172. (IF = 4.3,四区)
- W Tang, H Zheng, S Xu, P Li, L Zhan, X Luo, Z Dai, Q Wang, G Yu*. MMINP: A computational framework of microbe-Metabolite interactions-based metabolic profiles predictor based on the O2-PLS algorithm. Gut Microbes. 2023, 15(1):2223349. (IF = 12.2,一区)
- S Xu, L Zhan, W Tang, Q Wang, Z Dai, L Zhou, T Feng, M Chen, T Wu, E Hu, G Yu*. MicrobiotaProcess: A comprehensive R package for deep mining microbiome. The Innovation. 2023, 4(2):100388. (IF = 33.2,一区) (封面导读:Microbiome Toolkit)
- S Guo*, D Zhang, H Wang, Q An, G Yu, J Han, C Jiang, J Huang. Computational and Systematic Analysis of Multi-omics Data for Drug Discovery and Development. Frontiers in Medicine. 2023, 10:1146896. (IF = 3.1,三区)
2022 Link to heading
- S Xu, L Li, X Luo, M Chen, W Tang, L Zhan, Z Dai, TT. Lam, Y Guan, G Yu*. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta, 2022, 1(4):e56.
- Q Wang#, M Li#, T Wu, L Zhan, L Li, M Chen, W Xie, Z Xie, E Hu, S Xu, G Yu*. Exploring epigenomic datasets by ChIPseeker. Current Protocols, 2022, 2(10): e585.
- Z Dai, T Wu, S Xu, L Zhou, W Tang, E Hu, L Zhan M Chen, G Yu*. Characterization of toxin-antitoxin systems from public sequencing data: a case study in Pseudomonas aeruginosa. Frontiers in Microbiology. 2022, 13:951774. (IF = 5.2,二区,Top期刊)
- T Feng#, T Wu#, Y Zhang#, L Zhou#, S Liu, L Li, M Li, E Hu, Q Wang, X Fu, L Zhan, Z Xie, W Xie, X Huang*, X Shang*, G Yu*. Stemness Analysis Uncovers That The Peroxisome Proliferator-Activated Receptor Signaling Pathway Can Mediate Fatty Acid Homeostasis In Sorafenib-Resistant Hepatocellular Carcinoma Cells. Frontiers in Oncology. 2022, 12:912694. (IF = 4.7,三区)
- L Zhou#, T Feng#, S Xu, F Gao, TT Lam, Q Wang, T Wu, H Huang, L Zhan, L Li, Y Guan, Z Dai*, G Yu*. ggmsa: a visual exploration tool for multiple sequence alignment and associated data. Briefings in Bioinformatics. 2022, 23(4):bbac222. (IF = 9.5,一区,Top期刊)
- N Sato, Y Tamada, G Yu, Y Okuno*. CBNplot: Bayesian network plots for enrichment analysis. Bioinformatics. 2022, 38(10):2959-2960. (IF=5.8,一区,Top期刊)
- T Wu, CL Xiao, TT Lam, G Yu*. Biomedical Data Visualization: Methods and Applications. Frontiers in Genetics. 2022, 13:890775. (IF=3.7,三区)
- X Zhao#, W Tang#, H Wan#, Z Lan H Qin, Q Lin, Y Hu, G Yu*, N Jiang*, B Yu*. Altered gut microbiota as an auxiliary diagnostic indicator for patients with fracture-related infection. Frontiers in Microbiology. 2022, 13:723791. (IF = 5.2,二区,Top期刊)
2021 Link to heading
- S Xu#, M Chen#, T Feng, L Zhan, L Zhou, G Yu*. Use ggbreak to effectively utilize plotting space to deal with large datasets and outliers. Frontiers in Genetics. 2021, 12:774846. (IF=4.772,三区)
- CH Gao, G Yu, P Cai*. ggVennDiagram: An intuitive, easy-to-use, and highly customizable R package to generate Venn diagram. Frontiers in Genetics. 2021, 12:706907. (IF=4.772,三区)
- S Xu, Z Dai, P Guo, X Fu, S Liu, L Zhou, W Tang, T Feng, M Chen, L Zhan, T Wu, E Hu, Y Jiang*, X Bo*, G Yu*. ggtreeExtra: Compact visualization of richly annotated phylogenetic data. Molecular Biology and Evolution. 2021, 38(9):4039-4042. (IF=8.800,一区,Top期刊)
- T Wu#, E Hu#, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo*, G Yu*. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021, 2(3):100141. (封面导读:ClusterProfiler 4: Pathway Discovery)
- D Zeng#, Z Ye#, R Shen, G Yu, J Wu, Y Xiong, R Zhou, W Qiu, N Huang, L Sun, X Li, J Bin, Y Liao, M Shi, W Liao*. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Frontiers in Immunology. 2021, 12:687975. (IF=8.786,二区,Top期刊)
- T Wu, E Hu, X Ge*, G Yu*. nCov2019: an R package for studying the COVID-19 coronavirus pandemic. PeerJ. 2021, 9:e11421. (IF=3.061,三区)
- R Huang, C Soneson, FGM Ernst, KC Rue-Albrecht, G Yu, SC Hicks, MD Robinson*. TreeSummarizedExperiment: a S4 class for data with hierarchical structure. F1000Research. 2021, 9:1246.
- S Liu, G Yu*, L Liu*, X Zou, L Zhou, E Hu, Y Song. Identification of prognostic Stromal-Immune Score-Based Genes in Hepatocellular Carcinoma microenvironment. Frontiers in Genetics. 2021, 12:625236. (IF=4.772,三区)
2020 Link to heading
- J He#, S Xu#, B Zhang, C Xiao, Z Chen, F Si, J Fu, X Lin, G Zheng*, G Yu*, J Chen*. Gut microbiota and metabolite alterations associated with reduced bone mineral density or bone metabolic indexes in postmenopausal osteoporosis. Aging (Albany NY). 2020, 12(9):8583-8604. (IF=5.682,二区)
- G Yu*. Using ggtree to Visualize Data on Tree-Like Structure. Current Protocols in Bioinformatics. 2020, 69(1):e96. (Invited paper)
- LG Wang, TTY Lam, S Xu, Z Dai, L Zhou, T Feng, P Guo, CW Dunn, BR Jones, T Bradley, H Zhu, Y Guan, Y Jiang, G Yu*. treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Molecular Biology and Evolution. 2020, 37(2):599-603. (IF=16.240,一区,Top期刊)
- Z Hao, D Lv, Y Ge, J Shi, D Weijers, G Yu*, J Chen*. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Computer Science. 2020, 6:e251. (IF=1.392,三区)
2018 Link to heading
- G Yu*, TTY Lam, H Zhu, Y Guan*. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular Biology and Evolution. 2018, 35(12):3041-3043. (IF=14.797,一区, Top期刊)
- G Yu*. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics. 2018, 34(21):3766-3767. (IF=4.531, 一区, Top期刊)
Selected publications (2010-2017) Link to heading
- G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2017, 8(1):28-36. (IF=6.363, 一区,Top期刊)
- G Yu, QY He*. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Molecular BioSystems. 2016, 12(2):477-479. (IF=2.781,三区)
- G Yu*, LG Wang, QY He*. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparision and visualization. Bioinformatics. 2015, 31(14):2382-2383. (IF=5.766, 一区, Top期刊)
- G Yu*, LG Wang, GR Yan, QY He*. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics. 2015, 31(4):608-609. (IF=5.766, 一区, Top期刊)
- G Yu, LG Wang, Y Han, QY He*. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287. (IF=2.730, 四区)
- G Yu#, F Li#, Y Qin, X Bo*, Y Wu, S Wang*. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010, 26(7):976-978. (IF=4.877, 二区, Top期刊)