Integrating Tree Data: Methods and Applications
We have proposed and developed a series of methods and software tools for the operation, integration, and visualization of phylogenetic trees and data. Key innovations include: (1) introducing graphic grammar to the field of phylogenetics for the first time; (2) enhancing the data integration capabilities of phylogenetics and its application across various disciplines; (3) proposing two universal methods for phylogenetic data integration and visualization; and (4) designing data structures that can store phylogenetic trees, associated data, and visualization directives to ensure analytical reproducibility. These methods and tools offer a concise and unified syntax system, assisting researchers in discovering hidden patterns and proposing new hypotheses by integrating heterogeneous data within the context of evolution or hierarchy.
Two monographs have been published to introduce this series of work: “Data integration, manipulation and visualization of phylogenetic trees” (in English) by CRC Press and 《R实战:系统发育树的数据集成操作与可视化》 (in Chinese) by Publishing House of Electronics Industry (电子工业出版社).
1. Frist-time implementation of graphical grammar for visualizing phylogenetic tree data Link to heading
|
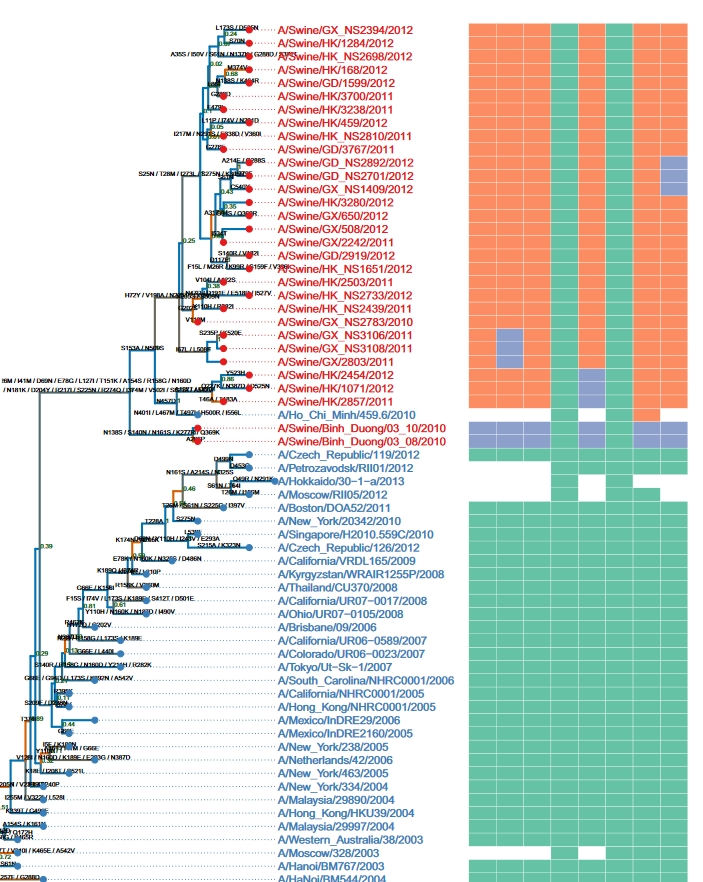
Numerous software tools exist for visualizing phylogenetic trees, but they primarily focus on displaying the tree’s topological structure and often lack comprehensive support for annotating the tree with additional knowledge or data. For the first time, ggtree introduces the grammar of graphics into the visualization of phylogenetic trees and related data. This innovative approach enables a high level of abstraction for visualization through a simple grammar, significantly reducing the complexity of data visualization and accommodating complex requirements. This work was published in Methods in Ecology and Evolution in 2017 (ESI highly cited) and was chosen by the journal as one of the ten representative works for its 10th anniversary celebration. An invited protocol paper demonstrating the use of this package was also published in Current Protocols in Bioinformatics in 2020. |
2. Base classes and functions for phylogenetic tree input and output Link to heading
|
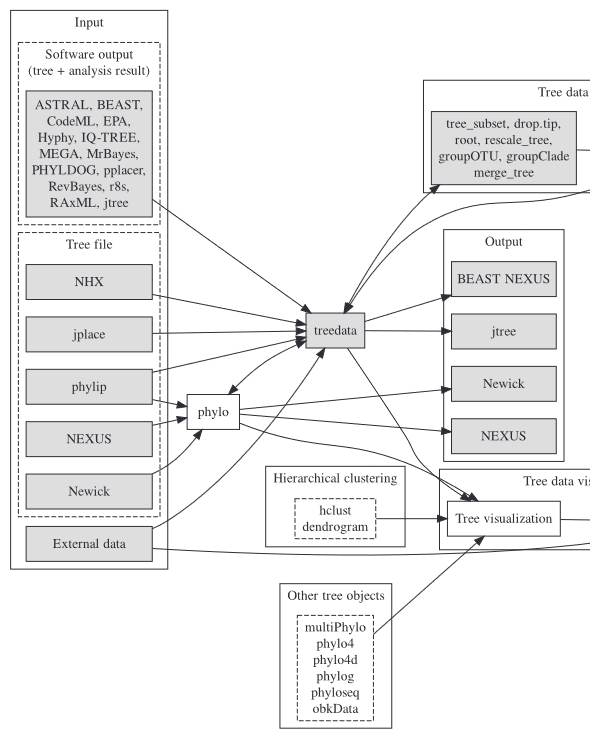
The outputs of phylogenetic software are often in non-standard formats, leading to compatibility issues and hindering integration and comparative analysis in downstream applications. To address this challenge, we developed treeio, a tool capable of parsing both standard and a variety of non-standard data formats. It facilitates the integration of external data and supports exporting phylogenetic trees and associated data into a single file. This dual capability enables data format conversion, thereby indirectly expanding the compatibility of software with a wider range of data. By parsing and integrating diverse data types, treeio empowers downstream integrated and comparative analyses, thus broadening the application scope of phylogenetic analysis. Our work, published in Molecular Biology and Evolution in 2020 (ESI highly cited), underscores the significance of this advancement. |
3. Proposing two general methods for the integration and visualization of phylogenetic data Link to heading
|
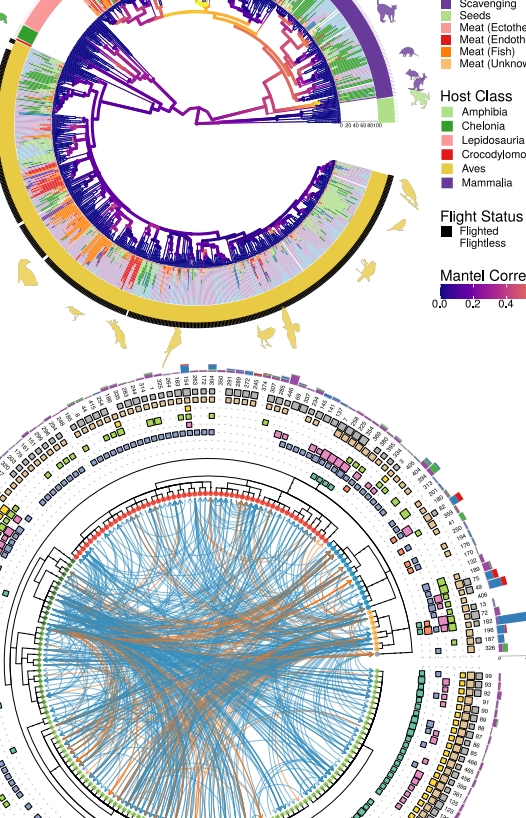
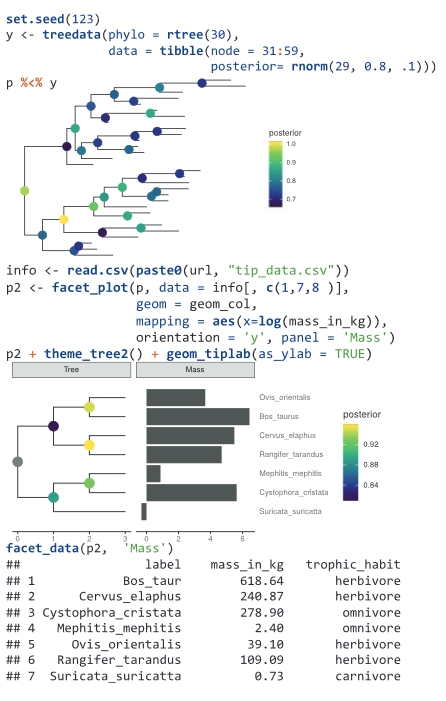
Two comprehensive methods have been proposed and implemented to address all facets of integrating and visualizing phylogenetic data. The first method enables data to be directly mapped onto the tree’s topology, facilitating the direct display of data or its transformation into visualization features. The second method involves the restructuring external data based on the tree’s topology, allowing users to visualize it according to their specifications, and subsequently aligning the visualization with the phylogenetic tree. These two methods enable the integration of diverse heterogeneous data from various disciplines within the context of phylogenetics, thereby aiding in the discovery of new patterns or the formulation of novel hypotheses. This work, published in Molecular Biology and Evolution in 2018 (ESI highly cited), served as the foundation for the development of the ggtreeExtra package. This package enhances integration and visualization capabilities for data richly annotated with additional features, and was published in Molecular Biology and Evolution in 2021. |
4. Enhancing data reuse and analytical reproducibility Link to heading
|
Visualisation of phylogenetic trees typically manifests as static images, leading to a lack of reusability of both the trees and associated data, thus impeding the integration of phylogenetic knowledge and comparative analysis. Research indicates that approximately 60% of published phylogenetic data is lost permanently. To tackle this issue, we devised the ggtree object, which encapsulates the phylogenetic tree, data, and visualization directives. This object can be rendered into an image, from which the phylogenetic tree and related data can be extracted. Furthermore, akin to the “format painter” tool, the visualization directives are transferable for visualizing other tree objects. Our efforts, published in iMeta in 2022, promote data reusability and research replicability, while facilitating the integration and comparative analysis of phylogenetic data within the field. |
5. Expanding support to other tree-like structures Link to heading
|
Broaden the scope of tools pertaining to tree data integration and visualization to encompass other tree-like structures, such as hierarchical clustering and classification/regression trees. We have developed the ggtreeDendro package to accommodate general hierarchical structures and are currently working on the ecluster package to support various omics data structures available in the Bioconductor project. These advancements enable the interpretation and integration of related data at the feature or sample level based on their hierarchical relationships. |
Feedback from the academic community Link to heading
|

|
Publications Link to heading
- M Chen#, X Luo#, S Xu#, L Li, J Li, Z Xie, Q Wang, Y Liao, B Liu, W Liang, K Mo, Q Song, X Chen*, TTY Lam*, G Yu*. Scalable method for exploring phylogenetic placement uncertainty with custom visualizations using treeio and ggtree. iMeta. 2025, 4(1):e269.
- L Zhan#, X Luo#, W Xie#, XA Zhu#, Z Xie, J Lin, L Li, W Tang, R Wang, L Deng, Y Liao, B Liu, Y Cai, Q Wang, S Xu*, G Yu*. shinyTempSignal: an R shiny application for exploring temporal and other phylogenetic signals. Journal of Genetics and Genomics. 2024, 51(7):762-768.
- L Li, W Xie, L Zhan, S Wen, X Luo, S Xu, Y Cai, W Tang, Q Wang, M Li, Z Xie, L Deng, H Zhu, G Yu*. Resolving Tumor Evolution: A Phylogenetic Approach. Journal of the National Cancer Center. 2024, 4(2):97-106.
- S Xu, L Li, X Luo, M Chen, W Tang, L Zhan, Z Dai, TT. Lam, Y Guan, G Yu*. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta, 2022, 1(4):e56.
- G Yu. Data Integration, Manipulation and Visualization of Phylogenetic Treess (1st edition). Chapman and Hall/CRC, 2022. doi: 10.1201/9781003279242
- L Zhou#, T Feng#, S Xu, F Gao, TT Lam, Q Wang, T Wu, H Huang, L Zhan, L Li, Y Guan, Z Dai*, G Yu*. ggmsa: a visual exploration tool for multiple sequence alignment and associated data. Briefings in Bioinformatics. 2022, 23(4):bbac222.
- S Xu, Z Dai, P Guo, X Fu, S Liu, L Zhou, W Tang, T Feng, M Chen, L Zhan, T Wu, E Hu, Y Jiang*, X Bo*, G Yu*. ggtreeExtra: Compact visualization of richly annotated phylogenetic data. Molecular Biology and Evolution. 2021, 38(9):4039-4042.
- G Yu*. Using ggtree to Visualize Data on Tree-Like Structure. Current Protocols in Bioinformatics. 2020, 69(1):e96.
- LG Wang, TTY Lam, S Xu, Z Dai, L Zhou, T Feng, P Guo, CW Dunn, BR Jones, T Bradley, H Zhu, Y Guan, Y Jiang, G Yu*. treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Molecular Biology and Evolution. 2020, 37(2):599-603.
- G Yu*, TTY Lam, H Zhu, Y Guan*. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular Biology and Evolution. 2018, 35(12):3041-3043.
- G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2017, 8(1):28-36.