5 Phylogenetic Tree Annotation
5.1 Visualizing and Annotating Tree Using Grammar of Graphics
The ggtree (Yu et al., 2017) is designed for a more general-purpose or a specific type of tree visualization and annotation. It supports the grammar of graphics implemented in ggplot2 and users can freely visualize/annotate a tree by combining several annotation layers.
library(ggtree)
treetext = "(((ADH2:0.1[&&NHX:S=human], ADH1:0.11[&&NHX:S=human]):
0.05 [&&NHX:S=primates:D=Y:B=100],ADHY:
0.1[&&NHX:S=nematode],ADHX:0.12 [&&NHX:S=insect]):
0.1[&&NHX:S=metazoa:D=N],(ADH4:0.09[&&NHX:S=yeast],
ADH3:0.13[&&NHX:S=yeast], ADH2:0.12[&&NHX:S=yeast],
ADH1:0.11[&&NHX:S=yeast]):0.1[&&NHX:S=Fungi])[&&NHX:D=N];"
tree <- read.nhx(textConnection(treetext))
ggtree(tree) + geom_tiplab() +
geom_label(aes(x=branch, label=S), fill='lightgreen') +
geom_label(aes(label=D), fill='steelblue') +
geom_text(aes(label=B), hjust=-.5)
FIGURE 5.1: Annotating tree using the grammar of graphics. The NHX tree was annotated using the grammar of graphic syntax by combining different layers using the + operator. Species information was labeled in the middle of the branches. Duplication events were shown on the most recent common ancestor and clade bootstrap values were displayed near to it.
Here, as an example, we visualized the tree with several layers to display annotation stored in NHX tags, including a layer of geom_tiplab() to display tip labels (gene name in this case), a layer using geom_label() to show species information (S tag) colored by light green, a layer of duplication event information (D tag) colored by steelblue and another layer using geom_text() to show bootstrap value (B tag).
Layers defined in ggplot2 can be applied to ggtree directly as demonstrated in Figure 5.1 of using geom_label() and geom_text(). But ggplot2 does not provide graphic layers that are specifically designed for phylogenetic tree annotation. For instance, layers for tip labels, tree branch scale legend, highlight, or labeling clade are all unavailable. To make tree annotation more flexible, several layers have been implemented in ggtree (Table 5.1), enabling different ways of annotation on various parts/components of a phylogenetic tree.
| Layer | Description |
|---|---|
| geom_balance | Highlights the two direct descendant clades of an internal node |
| geom_cladelab | Annotates a clade with bar and text label (or image) |
| geom_facet | Plots associated data in a specific panel (facet) and aligns the plot with the tree |
| geom_hilight | Highlights selected clade with rectangular or round shape |
| geom_inset | Adds insets (subplots) to tree nodes |
| geom_label2 | The modified version of geom_label, with subset aesthetic supported |
| geom_nodepoint | Annotates internal nodes with symbolic points |
| geom_point2 | The modified version of geom_point, with subset aesthetic supported |
| geom_range | Bar layer to present uncertainty of evolutionary inference |
| geom_rootpoint | Annotates root node with symbolic point |
| geom_rootedge | Adds root-edge to a tree |
| geom_segment2 | The modified version of geom_segment, with subset aesthetic supported |
| geom_strip | Annotates associated taxa with bar and (optional) text label |
| geom_taxalink | Links related taxa |
| geom_text2 | The modified version of geom_text, with subset aesthetic supported |
| geom_tiplab | The layer of tip labels |
| geom_tippoint | Annotates external nodes with symbolic points |
| geom_tree | Tree structure layer, with multiple layouts supported |
| geom_treescale | Tree branch scale legend |
5.2 Layers for Tree Annotation
5.2.1 Colored strips
The ggtree (Yu et al., 2017) implements geom_cladelab() layer to annotate a selected clade with a bar indicating the clade with a corresponding label.
The geom_cladelab() layer accepts a selected internal node number and labels the corresponding clade automatically (Figure 5.2A). To get the internal node number, please refer to Chapter 2.
set.seed(2015-12-21)
tree <- rtree(30)
p <- ggtree(tree) + xlim(NA, 8)
p + geom_cladelab(node=45, label="test label") +
geom_cladelab(node=34, label="another clade")Users can set the parameter, align = TRUE, to align the clade label, offset, to adjust the position and color to set the color of the bar and label text, etc. (Figure 5.2B).
p + geom_cladelab(node=45, label="test label", align=TRUE,
offset = .2, textcolor='red', barcolor='red') +
geom_cladelab(node=34, label="another clade", align=TRUE,
offset = .2, textcolor='blue', barcolor='blue')Users can change the angle of the clade label text and relative position from text to bar via the parameter offset.text. The size of the bar and text can be changed via the parameters barsize and fontsize, respectively (Figure 5.2C).
p + geom_cladelab(node=45, label="test label", align=TRUE, angle=270,
hjust='center', offset.text=.5, barsize=1.5, fontsize=8) +
geom_cladelab(node=34, label="another clade", align=TRUE, angle=45)Users can also use geom_label() to label the text and can set the background color by fill parameter (Figure 5.2D).
p + geom_cladelab(node=34, label="another clade", align=TRUE,
geom='label', fill='lightblue')
FIGURE 5.2: Labeling clades. Default (A); aligning and coloring clade bar and text (B); changing size and angle (C) and using geom_label() with background color in the text (D).
In addition, geom_cladelab() allows users to use the image or phylopic to annotate the clades, and supports using aesthetic mapping to automatically annotate the clade with bar and text label or image (e.g., mapping variable to color the clade labels) (Figure 5.3).
dat <- data.frame(node = c(45, 34),
name = c("test label", "another clade"))
# The node and label is required when geom="text"
## or geom="label" or geom="shadowtext".
p1 <- p + geom_cladelab(data = dat,
mapping = aes(node = node, label = name, color = name),
fontsize = 3)
dt <- data.frame(node = c(45, 34),
image = c("7fb9bea8-e758-4986-afb2-95a2c3bf983d",
"0174801d-15a6-4668-bfe0-4c421fbe51e8"),
name = c("specie A", "specie B"))
# when geom="phylopic" or geom="image", the image of aes is required.
p2 <- p + geom_cladelab(data = dt,
mapping = aes(node = node, label = name, image = image),
geom = "phylopic", imagecolor = "black",
offset=1, offset.text=0.5)
# The color or size of image also can be mapped.
p3 <- p + geom_cladelab(data = dt,
mapping = aes(node = node, label = name,
image = image, color = name),
geom = "phylopic", offset = 1, offset.text=0.5)FIGURE 5.3: Labeling clades using aesthetic mapping. The geom_cladelab() layer allows users to use aesthetic mapping to annotate the clades (A); it supports using images or phylopic to annotate clades (B); mapping variable to change color or size of the text or image is also supported (C).
The geom_cladelab() layer also supports unrooted tree layouts (Figure 5.4A).
ggtree(tree, layout="daylight") +
geom_cladelab(node=35, label="test label", angle=0,
fontsize=8, offset=.5, vjust=.5) +
geom_cladelab(node=55, label='another clade',
angle=-95, hjust=.5, fontsize=8)The geom_cladelab() is designed for labeling Monophyletic (Clade) while there are related taxa that do not form a clade. In ggtree, we provide another layer, geom_strip(), to add a strip/bar to indicate the association with an optional label for Polyphyletic or Paraphyletic (Figure 5.4B).
p + geom_tiplab() +
geom_strip('t10', 't30', barsize=2, color='red',
label="associated taxa", offset.text=.1) +
geom_strip('t1', 't18', barsize=2, color='blue',
label = "another label", offset.text=.1)
FIGURE 5.4: Labeling associated taxa. The geom_cladelab() is designed for labeling Monophyletic and supports unrooted layouts (A). The geom_strip() is designed for labeling all types of associated taxa, including Monophyletic, Polyphyletic, and Paraphyletic (B).
5.2.2 Highlight clades
The ggtree implements the geom_hilight() layer, which accepts an internal node number and adds a layer of a rectangle to highlight the selected clade (Figure 5.5)10.
nwk <- system.file("extdata", "sample.nwk", package="treeio")
tree <- read.tree(nwk)
ggtree(tree) +
geom_hilight(node=21, fill="steelblue", alpha=.6) +
geom_hilight(node=17, fill="darkgreen", alpha=.6)
ggtree(tree, layout="circular") +
geom_hilight(node=21, fill="steelblue", alpha=.6) +
geom_hilight(node=23, fill="darkgreen", alpha=.6)The geom_hilight layer also supports highlighting clades for unrooted layout trees with round (‘encircle’) or rectangular (‘rect’) shape (Figure 5.5C).
## type can be 'encircle' or 'rect'
pg + geom_hilight(node=55, linetype = 3) +
geom_hilight(node=35, fill='darkgreen', type="rect")Another way to highlight selected clades is by setting the clades with different colors and/or line types as demonstrated in Figure 6.2.
In addition to geom_hilight(), ggtree also implements geom_balance()
which is designed to highlight neighboring subclades of a given internal node (Figure 5.5D).
ggtree(tree) +
geom_balance(node=16, fill='steelblue', color='white', alpha=0.6, extend=1) +
geom_balance(node=19, fill='darkgreen', color='white', alpha=0.6, extend=1) The geom_hilight() layer supports using aesthetic mapping to automatically highlight clades as demonstrated in Figures 5.5E-F. For plot in Cartesian coordinates (e.g., rectangular layout), the rectangle can be rounded (Figure 5.5E) or filled with gradient colors (Figure 5.5F).
## using external data
d <- data.frame(node=c(17, 21), type=c("A", "B"))
ggtree(tree) + geom_hilight(data=d, aes(node=node, fill=type),
type = "roundrect")
## using data stored in the tree object
x <- read.nhx(system.file("extdata/NHX/ADH.nhx", package="treeio"))
ggtree(x) + geom_hilight(mapping=aes(subset = node %in% c(10, 12),
fill = S),
type = "gradient", gradient.direction = 'rt',
alpha = .8) +
scale_fill_manual(values=c("steelblue", "darkgreen"))FIGURE 5.5: Highlight selected clades. Rectangular layout (A), circular/fan (B), and unrooted layouts. Highlight neighboring subclades simultaneously (D). Highlight selected clades using associated data (E and F).
5.2.3 Taxa connection
Some evolutionary events (e.g., reassortment, horizontal gene transfer) cannot be modeled by a simple tree. The ggtree provides the geom_taxalink() layer that allows drawing straight or curved lines between any of two nodes in the tree, allowing it to represent evolutionary events by connecting taxa. It works with rectangular (Figure 5.6A), circular (Figure 5.6B), and inward circular (Figure 5.6C) layouts. The geom_taxalink() is not only useful for presenting evolutionary events, but it can also be used to combine evolutionary trees to present relationships or interactions between species (Xu, Dai, et al., 2021).
The geom_taxalink() layout supports aesthetic mapping, which requires a data.frame that stores association information with/without meta-data as input (Figure 5.6D).
p1 <- ggtree(tree) + geom_tiplab() + geom_taxalink(taxa1='A', taxa2='E') +
geom_taxalink(taxa1='F', taxa2='K', color='red', linetype = 'dashed',
arrow=arrow(length=unit(0.02, "npc")))
p2 <- ggtree(tree, layout="circular") +
geom_taxalink(taxa1='A', taxa2='E', color="grey", alpha=0.5,
offset=0.05, arrow=arrow(length=unit(0.01, "npc"))) +
geom_taxalink(taxa1='F', taxa2='K', color='red',
linetype = 'dashed', alpha=0.5, offset=0.05,
arrow=arrow(length=unit(0.01, "npc"))) +
geom_taxalink(taxa1="L", taxa2="M", color="blue", alpha=0.5,
offset=0.05, hratio=0.8,
arrow=arrow(length=unit(0.01, "npc"))) +
geom_tiplab()
# when the tree was created using reverse x,
# we can set outward to FALSE, which will generate the inward curve lines.
p3 <- ggtree(tree, layout="inward_circular", xlim=150) +
geom_taxalink(taxa1='A', taxa2='E', color="grey", alpha=0.5,
offset=-0.2, outward=FALSE,
arrow=arrow(length=unit(0.01, "npc"))) +
geom_taxalink(taxa1='F', taxa2='K', color='red', linetype = 'dashed',
alpha=0.5, offset=-0.2, outward=FALSE,
arrow=arrow(length=unit(0.01, "npc"))) +
geom_taxalink(taxa1="L", taxa2="M", color="blue", alpha=0.5,
offset=-0.2, outward=FALSE,
arrow=arrow(length=unit(0.01, "npc"))) +
geom_tiplab(hjust=1)
dat <- data.frame(from=c("A", "F", "L"),
to=c("E", "K", "M"),
h=c(1, 1, 0.1),
type=c("t1", "t2", "t3"),
s=c(2, 1, 2))
p4 <- ggtree(tree, layout="inward_circular", xlim=c(150, 0)) +
geom_taxalink(data=dat,
mapping=aes(taxa1=from,
taxa2=to,
color=type,
size=s),
ncp=10,
offset=0.15) +
geom_tiplab(hjust=1) +
scale_size_continuous(range=c(1,3))
plot_list(p1, p2, p3, p4, ncol=2, tag_levels='A')FIGURE 5.6: Linking related taxa. This can be used to indicate evolutionary events or relationships between species. Rectangular layout (A), circular layout (B), and inward circular layout (C and D). It supports aesthetic mapping to map variables to set line sizes and colors (D).
5.2.4 Uncertainty of evolutionary inference
The geom_range() layer supports displaying interval (highest posterior density, confidence interval, range) as horizontal bars on tree nodes. The center of the interval will anchor to the corresponding node. The center by default is the mean value of the interval (Figure 5.7A). We can set the center to the estimated mean or median value (Figure 5.7B), or the observed value. As the tree branch and the interval may not be on the same scale, ggtree provides scale_x_range to add a second x-axis for the range (Figure 5.7C). Note that x-axis is disabled by the default theme, and we need to enable it if we want to display it (e.g., using theme_tree2()).
file <- system.file("extdata/MEGA7", "mtCDNA_timetree.nex", package = "treeio")
x <- read.mega(file)
p1 <- ggtree(x) + geom_range('reltime_0.95_CI', color='red', size=3, alpha=.3)
p2 <- ggtree(x) + geom_range('reltime_0.95_CI', color='red', size=3,
alpha=.3, center='reltime')
p3 <- p2 + scale_x_range() + theme_tree2()FIGURE 5.7: Displaying uncertainty of evolutionary inference. The center (mean value of the range (A) or estimated value (B)) is anchored to the tree nodes. A second x-axis was used for range scaling (C).
5.3 Tree Annotation with Output from Evolution Software
5.3.1 Tree annotation using data from evolutionary analysis software
Chapter 1 introduced using treeio package (Wang et al., 2020) to parse different tree formats and commonly used software outputs to obtain phylogeny-associated data. These imported data, as S4 objects, can be visualized directly using ggtree. Figure 5.1 demonstrates a tree annotated using the information (species classification, duplication event, and bootstrap value) stored in the NHX file. PHYLDOG and pkg_revbayes output NHX files that can be parsed by treeio and visualized by ggtree with annotation using their inference data.
Furthermore, the evolutionary data from the inference of BEAST, MrBayes, and RevBayes, dN/dS values inferred by CODEML, ancestral sequences inferred by HyPhy, CODEML, or BASEML and short read placement by EPA and PPLACER can be used to annotate the tree directly.
file <- system.file("extdata/BEAST", "beast_mcc.tree", package="treeio")
beast <- read.beast(file)
ggtree(beast, aes(color=rate)) +
geom_range(range='length_0.95_HPD', color='red', alpha=.6, size=2) +
geom_nodelab(aes(x=branch, label=round(posterior, 2)), vjust=-.5, size=3) +
scale_color_continuous(low="darkgreen", high="red") +
theme(legend.position=c(.1, .8))FIGURE 5.8: Annotating BEAST tree with length_95%_HPD and posterior. Branch length credible intervals (95% HPD) were displayed as red horizontal bars and clade posterior values were shown on the middle of branches.
In Figure 5.8, the tree was visualized and annotated with posterior >0.9 and demonstrated length uncertainty (95% Highest Posterior Density (HPD) interval).
Ancestral sequences inferred by HyPhy can be parsed using treeio, whereas the substitutions along each tree branch were automatically computed and stored inside the phylogenetic tree object (i.e., S4 object). The ggtree package can utilize this information stored in the object to annotate the tree, as demonstrated in Figure 5.9.
nwk <- system.file("extdata/HYPHY", "labelledtree.tree",
package="treeio")
ancseq <- system.file("extdata/HYPHY", "ancseq.nex",
package="treeio")
tipfas <- system.file("extdata", "pa.fas", package="treeio")
hy <- read.hyphy(nwk, ancseq, tipfas)
ggtree(hy) +
geom_text(aes(x=branch, label=AA_subs), size=2,
vjust=-.3, color="firebrick")FIGURE 5.9: Annotating tree with amino acid substitution determined by ancestral sequences inferred by HyPhy. Amino acid substitutions were displayed in the middle of branches.
PAML’s BASEML and CODEML can also be used to infer ancestral sequences, whereas CODEML can infer selection pressure. After parsing this information using treeio, ggtree can integrate this information into the same tree structure and be used for annotation as illustrated in Figure 5.10.
rstfile <- system.file("extdata/PAML_Codeml", "rst",
package="treeio")
mlcfile <- system.file("extdata/PAML_Codeml", "mlc",
package="treeio")
ml <- read.codeml(rstfile, mlcfile)
ggtree(ml, aes(color=dN_vs_dS), branch.length='dN_vs_dS') +
scale_color_continuous(name='dN/dS', limits=c(0, 1.5),
oob=scales::squish,
low='darkgreen', high='red') +
geom_text(aes(x=branch, label=AA_subs),
vjust=-.5, color='steelblue', size=2) +
theme_tree2(legend.position=c(.9, .3))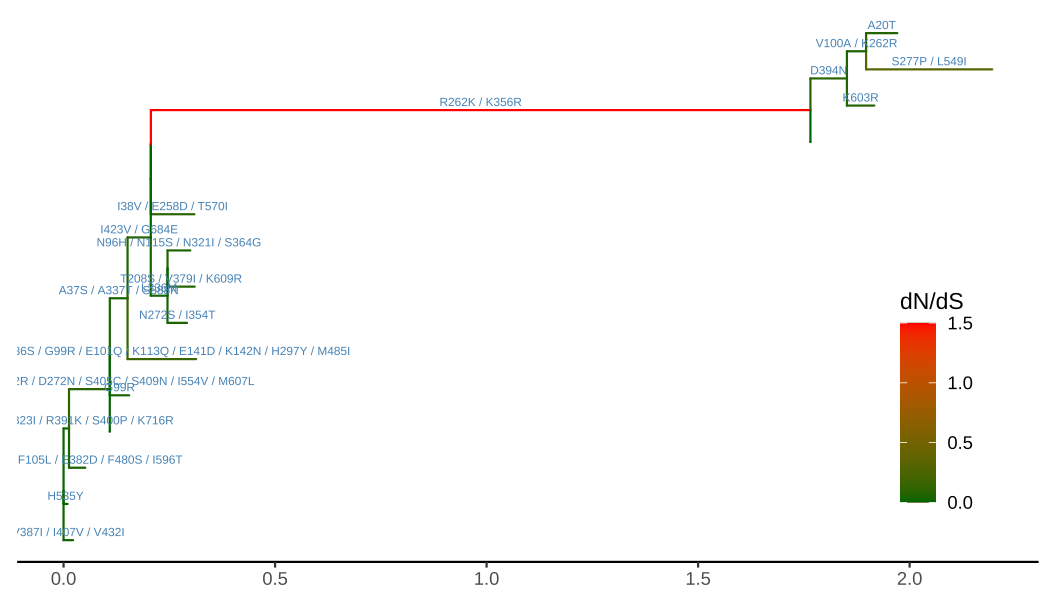
FIGURE 5.10: Annotating tree with amino acid substitution and dN/dS inferred by CODEML. Branches were rescaled and colored by dN/dS values, and amino acid substitutions were displayed on the middle of branches.
Not only all the tree data parsed by treeio can be used to visualize and annotate the phylogenetic tree using ggtree, but also other trees and tree-like objects defined in the R community are supported. The ggtree plays a unique role in the R ecosystem to facilitate phylogenetic analysis, and it can be easily integrated into other packages and pipelines. For more details of working with other tree-like structures, please refer to Chapter 9. In addition to direct support of tree objects, ggtree also allows users to plot a tree with different types of external data (see also Chapter 7 and (Yu et al., 2018)).
5.4 Summary
The ggtree package implements the grammar of graphics for annotating phylogenetic trees. Users can use the ggplot2 syntax to combine different annotation layers to produce complex tree annotation. If you are familiar with ggplot2, tree annotation with a high level of customization can be intuitive and flexible using ggtree. The ggtree can collect information in the treedata object or link external data to the structure of the tree. This will enable us to use the phylogenetic tree for data integration analysis and comparative studies, and will greatly expand the application of the phylogenetic tree in different fields.